
Lottiidibacillus patelloidae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Lottiidibacillus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
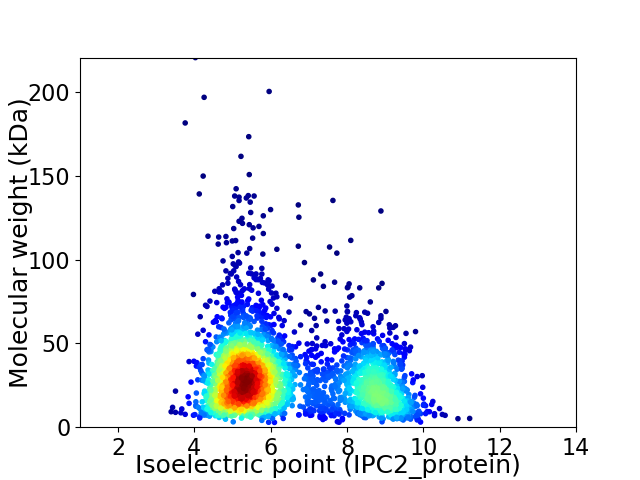
Virtual 2D-PAGE plot for 3053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A263BT11|A0A263BT11_9BACI SelB translation factor OS=Lottiidibacillus patelloidae OX=2670334 GN=selB PE=4 SV=1
MM1 pKa = 7.2KK2 pKa = 10.4RR3 pKa = 11.84KK4 pKa = 9.08LALFLTFILLFSQSVFPLNRR24 pKa = 11.84AFSEE28 pKa = 4.36YY29 pKa = 10.74GSNQIFSNVDD39 pKa = 3.0WLPYY43 pKa = 10.54AVGEE47 pKa = 4.49GNDD50 pKa = 4.46PLDD53 pKa = 3.78QNPNSTDD60 pKa = 2.86IAGNSEE66 pKa = 4.37LPAVSYY72 pKa = 10.85SYY74 pKa = 11.51DD75 pKa = 3.16STYY78 pKa = 11.03LYY80 pKa = 11.11FKK82 pKa = 10.14MRR84 pKa = 11.84LDD86 pKa = 4.7ANPLNNKK93 pKa = 9.18NDD95 pKa = 3.62NFDD98 pKa = 3.33QYY100 pKa = 11.94AWTVLFNTDD109 pKa = 3.17GNEE112 pKa = 4.05HH113 pKa = 6.53DD114 pKa = 3.76WEE116 pKa = 4.61YY117 pKa = 10.68MVSLNGLAEE126 pKa = 4.11TVQIGKK132 pKa = 8.33NTDD135 pKa = 3.16KK136 pKa = 11.19EE137 pKa = 4.08LDD139 pKa = 3.51SWGDD143 pKa = 3.52QAEE146 pKa = 4.29DD147 pKa = 3.71WLWTGPAIDD156 pKa = 4.55YY157 pKa = 11.21ADD159 pKa = 4.65ALPIDD164 pKa = 3.98VSLSNFNNDD173 pKa = 2.57QDD175 pKa = 4.28FFAYY179 pKa = 8.83WAVPWTDD186 pKa = 4.18FAAVTGLTPTSMIQLFYY203 pKa = 10.18ATSANASNYY212 pKa = 10.07NKK214 pKa = 10.29DD215 pKa = 3.21RR216 pKa = 11.84MGGHH220 pKa = 6.3PTSFADD226 pKa = 3.49AWGEE230 pKa = 3.78PILINGTTPNPPTIEE245 pKa = 3.94IAGGDD250 pKa = 3.62TKK252 pKa = 9.12TTNDD256 pKa = 3.68NTPLISGSSSAADD269 pKa = 3.63GSFVDD274 pKa = 3.41VTLYY278 pKa = 10.95LKK280 pKa = 10.52SDD282 pKa = 3.34PTKK285 pKa = 9.78TYY287 pKa = 9.41TYY289 pKa = 8.85QTTVEE294 pKa = 3.93NGQWQTLVTTALNNGVYY311 pKa = 9.74NVSAIVEE318 pKa = 4.03EE319 pKa = 4.87DD320 pKa = 3.56GQFGIDD326 pKa = 3.69EE327 pKa = 4.44QEE329 pKa = 4.14LTVDD333 pKa = 4.25PSQIIAPEE341 pKa = 3.88ITITTPSTGSAVTDD355 pKa = 3.51STPTISGSYY364 pKa = 9.7TSGTSTNNIVVEE376 pKa = 5.17FINGNNVVATGSASLANGVWEE397 pKa = 4.33FTPNSNLPLGTYY409 pKa = 7.15QVKK412 pKa = 8.46ATITDD417 pKa = 4.58EE418 pKa = 4.05INQTSTSSIEE428 pKa = 3.95LSVAEE433 pKa = 4.48AKK435 pKa = 10.32VLNINIDD442 pKa = 3.85TINPLVTNIVTPVFTGSSNAMDD464 pKa = 3.87GVTIFVTINNTSYY477 pKa = 7.77TTTVSNSSWSVQWPEE492 pKa = 3.56ALAEE496 pKa = 4.11GTYY499 pKa = 10.36VVSASVEE506 pKa = 4.29EE507 pKa = 4.21QGQTASDD514 pKa = 4.49SINLIVDD521 pKa = 3.56TTKK524 pKa = 9.5PTLSINGGTEE534 pKa = 3.49RR535 pKa = 11.84TTNEE539 pKa = 3.87STPTISGTTDD549 pKa = 2.93APDD552 pKa = 3.4NSVVTITLNGKK563 pKa = 9.86SYY565 pKa = 7.67TTTVTDD571 pKa = 3.85GTWTVDD577 pKa = 3.17VGEE580 pKa = 4.6LTDD583 pKa = 3.71GQYY586 pKa = 11.2EE587 pKa = 4.29VTATITDD594 pKa = 3.65EE595 pKa = 4.6AGNTTTAKK603 pKa = 10.12QSLTIDD609 pKa = 3.4TTGPAIVITSPEE621 pKa = 3.61NGAILNNHH629 pKa = 5.83TPTVTGTVEE638 pKa = 3.81AGSTVLVEE646 pKa = 5.89FIDD649 pKa = 3.91VNNSVVHH656 pKa = 6.25SGHH659 pKa = 5.97ATVTNEE665 pKa = 2.96NWTYY669 pKa = 10.9TPTGNLNEE677 pKa = 4.13GTYY680 pKa = 10.13SVKK683 pKa = 10.26ATGTDD688 pKa = 3.07ALGNPKK694 pKa = 8.84STSITITIDD703 pKa = 2.93VTAPTLTINGGTEE716 pKa = 3.59RR717 pKa = 11.84TTNDD721 pKa = 2.83STPTVSGTTDD731 pKa = 3.2APNGAIVTVTVNGQSYY747 pKa = 7.83TATVTDD753 pKa = 4.2GTWTVTVDD761 pKa = 3.27EE762 pKa = 5.87LIDD765 pKa = 3.42GTYY768 pKa = 10.61EE769 pKa = 3.8VTATITDD776 pKa = 3.48EE777 pKa = 4.48AGNVSTKK784 pKa = 8.89TQTLILSTIGPEE796 pKa = 4.05LKK798 pKa = 9.69IDD800 pKa = 3.88SPEE803 pKa = 3.96EE804 pKa = 3.75GALLSNYY811 pKa = 9.64HH812 pKa = 6.72PIISGTTNGDD822 pKa = 2.94KK823 pKa = 10.11VTITIYY829 pKa = 11.52DD830 pKa = 3.71MLEE833 pKa = 4.18KK834 pKa = 9.95IIEE837 pKa = 4.3TGVAEE842 pKa = 5.1LDD844 pKa = 3.48EE845 pKa = 5.29DD846 pKa = 5.31GNWTYY851 pKa = 10.5NTVSSLSDD859 pKa = 3.07GTYY862 pKa = 9.56LAKK865 pKa = 10.63AISTFNNGNDD875 pKa = 3.5TEE877 pKa = 4.58ASVSFIIDD885 pKa = 3.14VTAPTLTIDD894 pKa = 3.69GGTEE898 pKa = 3.51RR899 pKa = 11.84TTNDD903 pKa = 3.1STPTISGTTDD913 pKa = 2.93APDD916 pKa = 3.4NSVVTITLNGKK927 pKa = 9.86SYY929 pKa = 7.67TTTVTDD935 pKa = 4.05GTWTIDD941 pKa = 3.25TDD943 pKa = 3.67EE944 pKa = 4.77LTDD947 pKa = 3.44GTYY950 pKa = 10.84EE951 pKa = 3.98VTATITDD958 pKa = 3.61EE959 pKa = 4.61AGNSATAKK967 pKa = 10.04QSLTIDD973 pKa = 3.42TTGPTIAITSPEE985 pKa = 3.93NGAVLNDD992 pKa = 3.43QTPTVTGTVEE1002 pKa = 3.86AGSTVIVEE1010 pKa = 5.91FIDD1013 pKa = 3.63EE1014 pKa = 4.32NNSVVHH1020 pKa = 6.38SGHH1023 pKa = 5.9ATVANEE1029 pKa = 3.27NWTYY1033 pKa = 10.91TPTEE1037 pKa = 3.86NLLEE1041 pKa = 4.1GTYY1044 pKa = 7.93TVKK1047 pKa = 10.32ATGTDD1052 pKa = 3.09ALGNPKK1058 pKa = 8.84STSITITIDD1067 pKa = 2.93VTAPTLTIDD1076 pKa = 3.96GGAEE1080 pKa = 3.66RR1081 pKa = 11.84TTNDD1085 pKa = 2.82STPTVSGTTDD1095 pKa = 3.04APDD1098 pKa = 3.4NSVVTITLNGKK1109 pKa = 9.86SYY1111 pKa = 7.68TTTVTNGTWTIDD1123 pKa = 3.23VDD1125 pKa = 3.85EE1126 pKa = 5.03LTDD1129 pKa = 3.39GTYY1132 pKa = 10.84EE1133 pKa = 3.98VTATITDD1140 pKa = 3.53EE1141 pKa = 4.68AGNSTTATQSLTIDD1155 pKa = 3.4TLAPEE1160 pKa = 4.18VSIDD1164 pKa = 3.61NPLDD1168 pKa = 3.61NSVLEE1173 pKa = 4.93LGDD1176 pKa = 3.61IPITGTADD1184 pKa = 3.03VGSKK1188 pKa = 8.23VTLVLDD1194 pKa = 4.22DD1195 pKa = 4.88NEE1197 pKa = 5.5PITLTANEE1205 pKa = 4.07NGEE1208 pKa = 4.59WIYY1211 pKa = 11.03HH1212 pKa = 5.08PTPILSEE1219 pKa = 4.61GIHH1222 pKa = 5.89IVTVTAEE1229 pKa = 4.11DD1230 pKa = 3.67DD1231 pKa = 3.42QGNVSEE1237 pKa = 4.65PVSISFKK1244 pKa = 10.71VVDD1247 pKa = 4.42PNVIDD1252 pKa = 3.52EE1253 pKa = 4.52TEE1255 pKa = 3.86INITIDD1261 pKa = 5.39LSAQPQNILGNGIDD1275 pKa = 3.89HH1276 pKa = 7.48TILTAKK1282 pKa = 9.52LTDD1285 pKa = 3.75TNGEE1289 pKa = 3.92PLVNYY1294 pKa = 7.1EE1295 pKa = 4.54VKK1297 pKa = 10.64FSAPVGSFPEE1307 pKa = 4.24GFIAFTDD1314 pKa = 3.47QTGIATVKK1322 pKa = 10.01YY1323 pKa = 9.64VSAKK1327 pKa = 9.3TEE1329 pKa = 3.93SDD1331 pKa = 3.15EE1332 pKa = 4.23TLSYY1336 pKa = 10.66LVSAKK1341 pKa = 10.42AYY1343 pKa = 9.74NLDD1346 pKa = 3.63GNFVEE1351 pKa = 5.05QEE1353 pKa = 3.98QISISFNPAIISGVVIDD1370 pKa = 4.89NEE1372 pKa = 4.51TNEE1375 pKa = 4.94PIHH1378 pKa = 5.79NAKK1381 pKa = 9.97VVLSSDD1387 pKa = 4.11FNQDD1391 pKa = 2.45GTIDD1395 pKa = 3.61FTTTVLSDD1403 pKa = 3.39VEE1405 pKa = 4.3GKK1407 pKa = 10.31YY1408 pKa = 10.95VITIPNGNVSYY1419 pKa = 10.65LLEE1422 pKa = 3.84ITKK1425 pKa = 10.02PVLIGNTMQDD1435 pKa = 3.03VTFKK1439 pKa = 11.01QSVTAGSVSGSGTDD1453 pKa = 3.12EE1454 pKa = 4.64FYY1456 pKa = 10.28STEE1459 pKa = 4.04SATGIILLSDD1469 pKa = 4.22GSNGTLSLEE1478 pKa = 4.37NYY1480 pKa = 10.05DD1481 pKa = 4.08SIIVEE1486 pKa = 4.33VEE1488 pKa = 4.15DD1489 pKa = 3.91EE1490 pKa = 4.42SGDD1493 pKa = 3.47ITSLNGSVDD1502 pKa = 2.77ARR1504 pKa = 11.84TGVFLIDD1511 pKa = 3.57GLDD1514 pKa = 3.42VNKK1517 pKa = 10.64AYY1519 pKa = 8.87TLKK1522 pKa = 10.48FYY1524 pKa = 11.22YY1525 pKa = 10.57VFEE1528 pKa = 5.78DD1529 pKa = 4.09GSKK1532 pKa = 9.88IQVSSLAVVISNDD1545 pKa = 3.19GEE1547 pKa = 4.51LNISSCLIDD1556 pKa = 4.04PYY1558 pKa = 11.08GTITNKK1564 pKa = 8.52EE1565 pKa = 4.09TGEE1568 pKa = 4.24VIEE1571 pKa = 4.38GAVVEE1576 pKa = 5.23LYY1578 pKa = 10.95YY1579 pKa = 11.06ADD1581 pKa = 3.36TVRR1584 pKa = 11.84NQEE1587 pKa = 3.75NGRR1590 pKa = 11.84VKK1592 pKa = 10.24DD1593 pKa = 3.79TLVEE1597 pKa = 4.03LPILTFPPNDD1607 pKa = 2.97NHH1609 pKa = 6.23NPQNSSVEE1617 pKa = 3.87GKK1619 pKa = 9.08YY1620 pKa = 10.78AYY1622 pKa = 8.92MVYY1625 pKa = 10.44PEE1627 pKa = 4.36ADD1629 pKa = 3.41YY1630 pKa = 11.49YY1631 pKa = 10.73ILVKK1635 pKa = 10.84KK1636 pKa = 10.45NGYY1639 pKa = 7.78EE1640 pKa = 4.04LYY1642 pKa = 9.12EE1643 pKa = 4.45SPVIEE1648 pKa = 4.22VNQEE1652 pKa = 3.56IVRR1655 pKa = 11.84HH1656 pKa = 6.18DD1657 pKa = 3.71IEE1659 pKa = 4.71LTPIPQEE1666 pKa = 4.18LPATATSYY1674 pKa = 11.59YY1675 pKa = 9.69NYY1677 pKa = 10.47LVTGVLLLFIGVLTIFLYY1695 pKa = 10.51RR1696 pKa = 11.84RR1697 pKa = 11.84NRR1699 pKa = 11.84YY1700 pKa = 9.85SNN1702 pKa = 3.37
MM1 pKa = 7.2KK2 pKa = 10.4RR3 pKa = 11.84KK4 pKa = 9.08LALFLTFILLFSQSVFPLNRR24 pKa = 11.84AFSEE28 pKa = 4.36YY29 pKa = 10.74GSNQIFSNVDD39 pKa = 3.0WLPYY43 pKa = 10.54AVGEE47 pKa = 4.49GNDD50 pKa = 4.46PLDD53 pKa = 3.78QNPNSTDD60 pKa = 2.86IAGNSEE66 pKa = 4.37LPAVSYY72 pKa = 10.85SYY74 pKa = 11.51DD75 pKa = 3.16STYY78 pKa = 11.03LYY80 pKa = 11.11FKK82 pKa = 10.14MRR84 pKa = 11.84LDD86 pKa = 4.7ANPLNNKK93 pKa = 9.18NDD95 pKa = 3.62NFDD98 pKa = 3.33QYY100 pKa = 11.94AWTVLFNTDD109 pKa = 3.17GNEE112 pKa = 4.05HH113 pKa = 6.53DD114 pKa = 3.76WEE116 pKa = 4.61YY117 pKa = 10.68MVSLNGLAEE126 pKa = 4.11TVQIGKK132 pKa = 8.33NTDD135 pKa = 3.16KK136 pKa = 11.19EE137 pKa = 4.08LDD139 pKa = 3.51SWGDD143 pKa = 3.52QAEE146 pKa = 4.29DD147 pKa = 3.71WLWTGPAIDD156 pKa = 4.55YY157 pKa = 11.21ADD159 pKa = 4.65ALPIDD164 pKa = 3.98VSLSNFNNDD173 pKa = 2.57QDD175 pKa = 4.28FFAYY179 pKa = 8.83WAVPWTDD186 pKa = 4.18FAAVTGLTPTSMIQLFYY203 pKa = 10.18ATSANASNYY212 pKa = 10.07NKK214 pKa = 10.29DD215 pKa = 3.21RR216 pKa = 11.84MGGHH220 pKa = 6.3PTSFADD226 pKa = 3.49AWGEE230 pKa = 3.78PILINGTTPNPPTIEE245 pKa = 3.94IAGGDD250 pKa = 3.62TKK252 pKa = 9.12TTNDD256 pKa = 3.68NTPLISGSSSAADD269 pKa = 3.63GSFVDD274 pKa = 3.41VTLYY278 pKa = 10.95LKK280 pKa = 10.52SDD282 pKa = 3.34PTKK285 pKa = 9.78TYY287 pKa = 9.41TYY289 pKa = 8.85QTTVEE294 pKa = 3.93NGQWQTLVTTALNNGVYY311 pKa = 9.74NVSAIVEE318 pKa = 4.03EE319 pKa = 4.87DD320 pKa = 3.56GQFGIDD326 pKa = 3.69EE327 pKa = 4.44QEE329 pKa = 4.14LTVDD333 pKa = 4.25PSQIIAPEE341 pKa = 3.88ITITTPSTGSAVTDD355 pKa = 3.51STPTISGSYY364 pKa = 9.7TSGTSTNNIVVEE376 pKa = 5.17FINGNNVVATGSASLANGVWEE397 pKa = 4.33FTPNSNLPLGTYY409 pKa = 7.15QVKK412 pKa = 8.46ATITDD417 pKa = 4.58EE418 pKa = 4.05INQTSTSSIEE428 pKa = 3.95LSVAEE433 pKa = 4.48AKK435 pKa = 10.32VLNINIDD442 pKa = 3.85TINPLVTNIVTPVFTGSSNAMDD464 pKa = 3.87GVTIFVTINNTSYY477 pKa = 7.77TTTVSNSSWSVQWPEE492 pKa = 3.56ALAEE496 pKa = 4.11GTYY499 pKa = 10.36VVSASVEE506 pKa = 4.29EE507 pKa = 4.21QGQTASDD514 pKa = 4.49SINLIVDD521 pKa = 3.56TTKK524 pKa = 9.5PTLSINGGTEE534 pKa = 3.49RR535 pKa = 11.84TTNEE539 pKa = 3.87STPTISGTTDD549 pKa = 2.93APDD552 pKa = 3.4NSVVTITLNGKK563 pKa = 9.86SYY565 pKa = 7.67TTTVTDD571 pKa = 3.85GTWTVDD577 pKa = 3.17VGEE580 pKa = 4.6LTDD583 pKa = 3.71GQYY586 pKa = 11.2EE587 pKa = 4.29VTATITDD594 pKa = 3.65EE595 pKa = 4.6AGNTTTAKK603 pKa = 10.12QSLTIDD609 pKa = 3.4TTGPAIVITSPEE621 pKa = 3.61NGAILNNHH629 pKa = 5.83TPTVTGTVEE638 pKa = 3.81AGSTVLVEE646 pKa = 5.89FIDD649 pKa = 3.91VNNSVVHH656 pKa = 6.25SGHH659 pKa = 5.97ATVTNEE665 pKa = 2.96NWTYY669 pKa = 10.9TPTGNLNEE677 pKa = 4.13GTYY680 pKa = 10.13SVKK683 pKa = 10.26ATGTDD688 pKa = 3.07ALGNPKK694 pKa = 8.84STSITITIDD703 pKa = 2.93VTAPTLTINGGTEE716 pKa = 3.59RR717 pKa = 11.84TTNDD721 pKa = 2.83STPTVSGTTDD731 pKa = 3.2APNGAIVTVTVNGQSYY747 pKa = 7.83TATVTDD753 pKa = 4.2GTWTVTVDD761 pKa = 3.27EE762 pKa = 5.87LIDD765 pKa = 3.42GTYY768 pKa = 10.61EE769 pKa = 3.8VTATITDD776 pKa = 3.48EE777 pKa = 4.48AGNVSTKK784 pKa = 8.89TQTLILSTIGPEE796 pKa = 4.05LKK798 pKa = 9.69IDD800 pKa = 3.88SPEE803 pKa = 3.96EE804 pKa = 3.75GALLSNYY811 pKa = 9.64HH812 pKa = 6.72PIISGTTNGDD822 pKa = 2.94KK823 pKa = 10.11VTITIYY829 pKa = 11.52DD830 pKa = 3.71MLEE833 pKa = 4.18KK834 pKa = 9.95IIEE837 pKa = 4.3TGVAEE842 pKa = 5.1LDD844 pKa = 3.48EE845 pKa = 5.29DD846 pKa = 5.31GNWTYY851 pKa = 10.5NTVSSLSDD859 pKa = 3.07GTYY862 pKa = 9.56LAKK865 pKa = 10.63AISTFNNGNDD875 pKa = 3.5TEE877 pKa = 4.58ASVSFIIDD885 pKa = 3.14VTAPTLTIDD894 pKa = 3.69GGTEE898 pKa = 3.51RR899 pKa = 11.84TTNDD903 pKa = 3.1STPTISGTTDD913 pKa = 2.93APDD916 pKa = 3.4NSVVTITLNGKK927 pKa = 9.86SYY929 pKa = 7.67TTTVTDD935 pKa = 4.05GTWTIDD941 pKa = 3.25TDD943 pKa = 3.67EE944 pKa = 4.77LTDD947 pKa = 3.44GTYY950 pKa = 10.84EE951 pKa = 3.98VTATITDD958 pKa = 3.61EE959 pKa = 4.61AGNSATAKK967 pKa = 10.04QSLTIDD973 pKa = 3.42TTGPTIAITSPEE985 pKa = 3.93NGAVLNDD992 pKa = 3.43QTPTVTGTVEE1002 pKa = 3.86AGSTVIVEE1010 pKa = 5.91FIDD1013 pKa = 3.63EE1014 pKa = 4.32NNSVVHH1020 pKa = 6.38SGHH1023 pKa = 5.9ATVANEE1029 pKa = 3.27NWTYY1033 pKa = 10.91TPTEE1037 pKa = 3.86NLLEE1041 pKa = 4.1GTYY1044 pKa = 7.93TVKK1047 pKa = 10.32ATGTDD1052 pKa = 3.09ALGNPKK1058 pKa = 8.84STSITITIDD1067 pKa = 2.93VTAPTLTIDD1076 pKa = 3.96GGAEE1080 pKa = 3.66RR1081 pKa = 11.84TTNDD1085 pKa = 2.82STPTVSGTTDD1095 pKa = 3.04APDD1098 pKa = 3.4NSVVTITLNGKK1109 pKa = 9.86SYY1111 pKa = 7.68TTTVTNGTWTIDD1123 pKa = 3.23VDD1125 pKa = 3.85EE1126 pKa = 5.03LTDD1129 pKa = 3.39GTYY1132 pKa = 10.84EE1133 pKa = 3.98VTATITDD1140 pKa = 3.53EE1141 pKa = 4.68AGNSTTATQSLTIDD1155 pKa = 3.4TLAPEE1160 pKa = 4.18VSIDD1164 pKa = 3.61NPLDD1168 pKa = 3.61NSVLEE1173 pKa = 4.93LGDD1176 pKa = 3.61IPITGTADD1184 pKa = 3.03VGSKK1188 pKa = 8.23VTLVLDD1194 pKa = 4.22DD1195 pKa = 4.88NEE1197 pKa = 5.5PITLTANEE1205 pKa = 4.07NGEE1208 pKa = 4.59WIYY1211 pKa = 11.03HH1212 pKa = 5.08PTPILSEE1219 pKa = 4.61GIHH1222 pKa = 5.89IVTVTAEE1229 pKa = 4.11DD1230 pKa = 3.67DD1231 pKa = 3.42QGNVSEE1237 pKa = 4.65PVSISFKK1244 pKa = 10.71VVDD1247 pKa = 4.42PNVIDD1252 pKa = 3.52EE1253 pKa = 4.52TEE1255 pKa = 3.86INITIDD1261 pKa = 5.39LSAQPQNILGNGIDD1275 pKa = 3.89HH1276 pKa = 7.48TILTAKK1282 pKa = 9.52LTDD1285 pKa = 3.75TNGEE1289 pKa = 3.92PLVNYY1294 pKa = 7.1EE1295 pKa = 4.54VKK1297 pKa = 10.64FSAPVGSFPEE1307 pKa = 4.24GFIAFTDD1314 pKa = 3.47QTGIATVKK1322 pKa = 10.01YY1323 pKa = 9.64VSAKK1327 pKa = 9.3TEE1329 pKa = 3.93SDD1331 pKa = 3.15EE1332 pKa = 4.23TLSYY1336 pKa = 10.66LVSAKK1341 pKa = 10.42AYY1343 pKa = 9.74NLDD1346 pKa = 3.63GNFVEE1351 pKa = 5.05QEE1353 pKa = 3.98QISISFNPAIISGVVIDD1370 pKa = 4.89NEE1372 pKa = 4.51TNEE1375 pKa = 4.94PIHH1378 pKa = 5.79NAKK1381 pKa = 9.97VVLSSDD1387 pKa = 4.11FNQDD1391 pKa = 2.45GTIDD1395 pKa = 3.61FTTTVLSDD1403 pKa = 3.39VEE1405 pKa = 4.3GKK1407 pKa = 10.31YY1408 pKa = 10.95VITIPNGNVSYY1419 pKa = 10.65LLEE1422 pKa = 3.84ITKK1425 pKa = 10.02PVLIGNTMQDD1435 pKa = 3.03VTFKK1439 pKa = 11.01QSVTAGSVSGSGTDD1453 pKa = 3.12EE1454 pKa = 4.64FYY1456 pKa = 10.28STEE1459 pKa = 4.04SATGIILLSDD1469 pKa = 4.22GSNGTLSLEE1478 pKa = 4.37NYY1480 pKa = 10.05DD1481 pKa = 4.08SIIVEE1486 pKa = 4.33VEE1488 pKa = 4.15DD1489 pKa = 3.91EE1490 pKa = 4.42SGDD1493 pKa = 3.47ITSLNGSVDD1502 pKa = 2.77ARR1504 pKa = 11.84TGVFLIDD1511 pKa = 3.57GLDD1514 pKa = 3.42VNKK1517 pKa = 10.64AYY1519 pKa = 8.87TLKK1522 pKa = 10.48FYY1524 pKa = 11.22YY1525 pKa = 10.57VFEE1528 pKa = 5.78DD1529 pKa = 4.09GSKK1532 pKa = 9.88IQVSSLAVVISNDD1545 pKa = 3.19GEE1547 pKa = 4.51LNISSCLIDD1556 pKa = 4.04PYY1558 pKa = 11.08GTITNKK1564 pKa = 8.52EE1565 pKa = 4.09TGEE1568 pKa = 4.24VIEE1571 pKa = 4.38GAVVEE1576 pKa = 5.23LYY1578 pKa = 10.95YY1579 pKa = 11.06ADD1581 pKa = 3.36TVRR1584 pKa = 11.84NQEE1587 pKa = 3.75NGRR1590 pKa = 11.84VKK1592 pKa = 10.24DD1593 pKa = 3.79TLVEE1597 pKa = 4.03LPILTFPPNDD1607 pKa = 2.97NHH1609 pKa = 6.23NPQNSSVEE1617 pKa = 3.87GKK1619 pKa = 9.08YY1620 pKa = 10.78AYY1622 pKa = 8.92MVYY1625 pKa = 10.44PEE1627 pKa = 4.36ADD1629 pKa = 3.41YY1630 pKa = 11.49YY1631 pKa = 10.73ILVKK1635 pKa = 10.84KK1636 pKa = 10.45NGYY1639 pKa = 7.78EE1640 pKa = 4.04LYY1642 pKa = 9.12EE1643 pKa = 4.45SPVIEE1648 pKa = 4.22VNQEE1652 pKa = 3.56IVRR1655 pKa = 11.84HH1656 pKa = 6.18DD1657 pKa = 3.71IEE1659 pKa = 4.71LTPIPQEE1666 pKa = 4.18LPATATSYY1674 pKa = 11.59YY1675 pKa = 9.69NYY1677 pKa = 10.47LVTGVLLLFIGVLTIFLYY1695 pKa = 10.51RR1696 pKa = 11.84RR1697 pKa = 11.84NRR1699 pKa = 11.84YY1700 pKa = 9.85SNN1702 pKa = 3.37
Molecular weight: 181.66 kDa
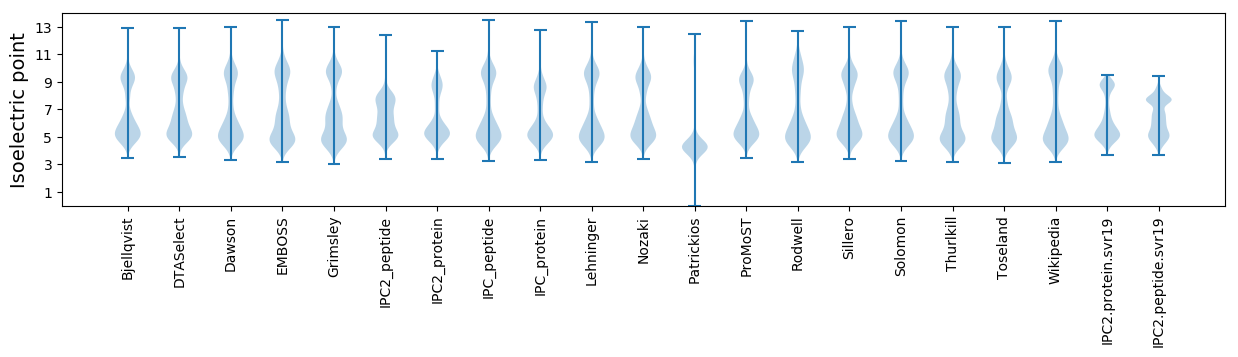
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A263BS86|A0A263BS86_9BACI Uncharacterized protein OS=Lottiidibacillus patelloidae OX=2670334 GN=CIB95_11750 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.02KK14 pKa = 8.47VHH16 pKa = 5.57GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTKK25 pKa = 10.29NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
875866 |
26 |
2026 |
286.9 |
32.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.496 ± 0.049 | 0.701 ± 0.013 |
5.066 ± 0.037 | 7.692 ± 0.051 |
4.697 ± 0.044 | 6.487 ± 0.046 |
2.052 ± 0.023 | 8.373 ± 0.044 |
7.528 ± 0.049 | 9.711 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.022 | 5.042 ± 0.035 |
3.356 ± 0.031 | 3.358 ± 0.027 |
3.719 ± 0.032 | 5.95 ± 0.033 |
5.502 ± 0.037 | 6.936 ± 0.04 |
0.977 ± 0.018 | 3.719 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
