
Kerstersia gyiorum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Kerstersia
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
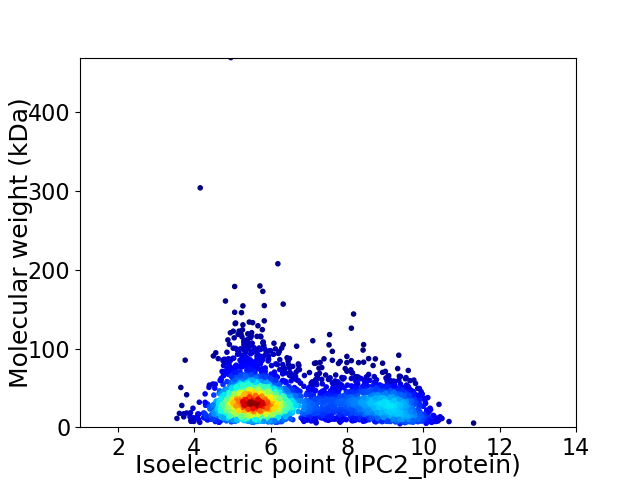
Virtual 2D-PAGE plot for 3267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
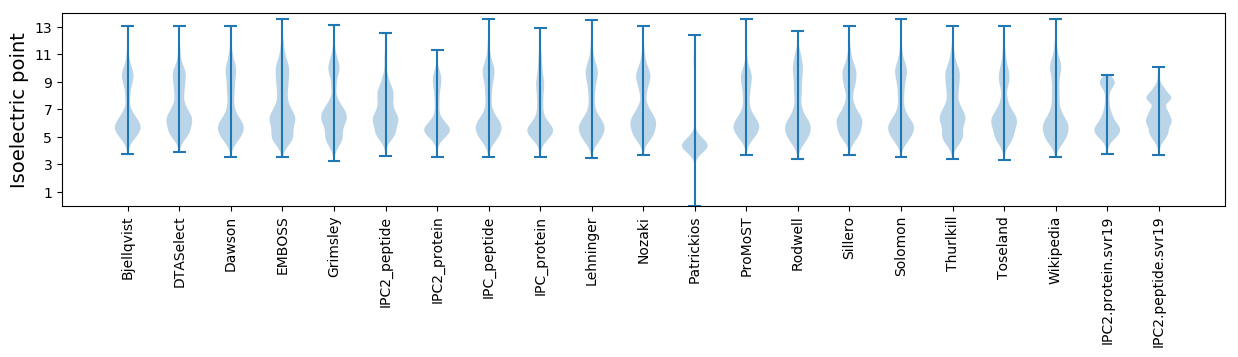
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A171KTL2|A0A171KTL2_9BURK Alkane 1-monooxygenase OS=Kerstersia gyiorum OX=206506 GN=AAV32_06985 PE=4 SV=1
MM1 pKa = 7.6GSYY4 pKa = 10.48KK5 pKa = 9.29VTPVSGNVLDD15 pKa = 4.53NDD17 pKa = 4.19AAGEE21 pKa = 4.26GAKK24 pKa = 8.75VTHH27 pKa = 7.1IDD29 pKa = 3.8GEE31 pKa = 4.6QVGANGTTTFEE42 pKa = 4.21GEE44 pKa = 4.31YY45 pKa = 11.05GSLTIGANGEE55 pKa = 4.22FTYY58 pKa = 10.5TPYY61 pKa = 9.88DD62 pKa = 3.68TNGLGIGKK70 pKa = 9.16VEE72 pKa = 4.07TFEE75 pKa = 4.11YY76 pKa = 9.61TIKK79 pKa = 10.86AADD82 pKa = 3.92GSEE85 pKa = 4.14STAEE89 pKa = 3.29IHH91 pKa = 6.92IRR93 pKa = 11.84IDD95 pKa = 3.06SDD97 pKa = 3.77GQGLIWPEE105 pKa = 4.6DD106 pKa = 3.56PSQPAEE112 pKa = 3.98VDD114 pKa = 3.71LVANNDD120 pKa = 3.43TGEE123 pKa = 4.25TVINSDD129 pKa = 3.83YY130 pKa = 11.08LVTQGGPSEE139 pKa = 4.09QAPSQTIGGIFAGAVTKK156 pKa = 10.23TSSVNFTVDD165 pKa = 3.47PDD167 pKa = 3.66SKK169 pKa = 11.62ADD171 pKa = 3.67VKK173 pKa = 10.16ITASSPDD180 pKa = 3.6EE181 pKa = 4.25LLVSDD186 pKa = 4.16SLKK189 pKa = 9.83VTVTGPNGYY198 pKa = 10.02SKK200 pKa = 9.65TYY202 pKa = 9.23TGTGGLFSNLSVQEE216 pKa = 3.94MLQGLEE222 pKa = 3.95AGNYY226 pKa = 7.16TVTATYY232 pKa = 10.13QRR234 pKa = 11.84SGTGTGGKK242 pKa = 9.91LEE244 pKa = 4.41LAYY247 pKa = 8.15EE248 pKa = 4.44TQSVTHH254 pKa = 6.59LDD256 pKa = 3.58EE257 pKa = 4.91YY258 pKa = 10.51VVKK261 pKa = 10.58DD262 pKa = 3.26IQAATGNVLADD273 pKa = 3.36NHH275 pKa = 6.83LGSTYY280 pKa = 10.72TKK282 pKa = 10.1FLVEE286 pKa = 4.18NGSGQFVAVANGTTVTGLYY305 pKa = 10.52GVLTINTDD313 pKa = 2.65GSYY316 pKa = 10.71SYY318 pKa = 11.66APTSSLAAIGKK329 pKa = 9.37EE330 pKa = 4.0DD331 pKa = 3.54VFTYY335 pKa = 10.57RR336 pKa = 11.84LEE338 pKa = 4.53HH339 pKa = 6.53PNGTVEE345 pKa = 4.28EE346 pKa = 4.3ATLTIAVEE354 pKa = 4.17HH355 pKa = 5.91GTGPFEE361 pKa = 5.36PDD363 pKa = 3.13ALDD366 pKa = 3.85SADD369 pKa = 4.02EE370 pKa = 4.28FSAFADD376 pKa = 3.25TDD378 pKa = 3.73AFEE381 pKa = 5.89DD382 pKa = 4.2EE383 pKa = 5.24SDD385 pKa = 3.8DD386 pKa = 4.0HH387 pKa = 8.41DD388 pKa = 5.78SDD390 pKa = 4.41VVALRR395 pKa = 11.84GLDD398 pKa = 3.76AEE400 pKa = 5.36DD401 pKa = 3.99GTDD404 pKa = 3.69AEE406 pKa = 4.75PATEE410 pKa = 4.03VAEE413 pKa = 4.7LNLDD417 pKa = 3.82DD418 pKa = 6.02LLYY421 pKa = 10.6EE422 pKa = 4.29PSEE425 pKa = 4.3EE426 pKa = 4.41GDD428 pKa = 3.29VDD430 pKa = 5.22LSALSEE436 pKa = 4.54DD437 pKa = 5.33GEE439 pKa = 4.39DD440 pKa = 3.93QEE442 pKa = 5.51DD443 pKa = 3.67AAAEE447 pKa = 4.01AQEE450 pKa = 4.51SVTAAVEE457 pKa = 3.96TDD459 pKa = 3.62EE460 pKa = 4.78PVVDD464 pKa = 4.08VPLVNPLDD472 pKa = 5.1DD473 pKa = 6.05DD474 pKa = 5.02LNQPQVFLGG483 pKa = 3.63
MM1 pKa = 7.6GSYY4 pKa = 10.48KK5 pKa = 9.29VTPVSGNVLDD15 pKa = 4.53NDD17 pKa = 4.19AAGEE21 pKa = 4.26GAKK24 pKa = 8.75VTHH27 pKa = 7.1IDD29 pKa = 3.8GEE31 pKa = 4.6QVGANGTTTFEE42 pKa = 4.21GEE44 pKa = 4.31YY45 pKa = 11.05GSLTIGANGEE55 pKa = 4.22FTYY58 pKa = 10.5TPYY61 pKa = 9.88DD62 pKa = 3.68TNGLGIGKK70 pKa = 9.16VEE72 pKa = 4.07TFEE75 pKa = 4.11YY76 pKa = 9.61TIKK79 pKa = 10.86AADD82 pKa = 3.92GSEE85 pKa = 4.14STAEE89 pKa = 3.29IHH91 pKa = 6.92IRR93 pKa = 11.84IDD95 pKa = 3.06SDD97 pKa = 3.77GQGLIWPEE105 pKa = 4.6DD106 pKa = 3.56PSQPAEE112 pKa = 3.98VDD114 pKa = 3.71LVANNDD120 pKa = 3.43TGEE123 pKa = 4.25TVINSDD129 pKa = 3.83YY130 pKa = 11.08LVTQGGPSEE139 pKa = 4.09QAPSQTIGGIFAGAVTKK156 pKa = 10.23TSSVNFTVDD165 pKa = 3.47PDD167 pKa = 3.66SKK169 pKa = 11.62ADD171 pKa = 3.67VKK173 pKa = 10.16ITASSPDD180 pKa = 3.6EE181 pKa = 4.25LLVSDD186 pKa = 4.16SLKK189 pKa = 9.83VTVTGPNGYY198 pKa = 10.02SKK200 pKa = 9.65TYY202 pKa = 9.23TGTGGLFSNLSVQEE216 pKa = 3.94MLQGLEE222 pKa = 3.95AGNYY226 pKa = 7.16TVTATYY232 pKa = 10.13QRR234 pKa = 11.84SGTGTGGKK242 pKa = 9.91LEE244 pKa = 4.41LAYY247 pKa = 8.15EE248 pKa = 4.44TQSVTHH254 pKa = 6.59LDD256 pKa = 3.58EE257 pKa = 4.91YY258 pKa = 10.51VVKK261 pKa = 10.58DD262 pKa = 3.26IQAATGNVLADD273 pKa = 3.36NHH275 pKa = 6.83LGSTYY280 pKa = 10.72TKK282 pKa = 10.1FLVEE286 pKa = 4.18NGSGQFVAVANGTTVTGLYY305 pKa = 10.52GVLTINTDD313 pKa = 2.65GSYY316 pKa = 10.71SYY318 pKa = 11.66APTSSLAAIGKK329 pKa = 9.37EE330 pKa = 4.0DD331 pKa = 3.54VFTYY335 pKa = 10.57RR336 pKa = 11.84LEE338 pKa = 4.53HH339 pKa = 6.53PNGTVEE345 pKa = 4.28EE346 pKa = 4.3ATLTIAVEE354 pKa = 4.17HH355 pKa = 5.91GTGPFEE361 pKa = 5.36PDD363 pKa = 3.13ALDD366 pKa = 3.85SADD369 pKa = 4.02EE370 pKa = 4.28FSAFADD376 pKa = 3.25TDD378 pKa = 3.73AFEE381 pKa = 5.89DD382 pKa = 4.2EE383 pKa = 5.24SDD385 pKa = 3.8DD386 pKa = 4.0HH387 pKa = 8.41DD388 pKa = 5.78SDD390 pKa = 4.41VVALRR395 pKa = 11.84GLDD398 pKa = 3.76AEE400 pKa = 5.36DD401 pKa = 3.99GTDD404 pKa = 3.69AEE406 pKa = 4.75PATEE410 pKa = 4.03VAEE413 pKa = 4.7LNLDD417 pKa = 3.82DD418 pKa = 6.02LLYY421 pKa = 10.6EE422 pKa = 4.29PSEE425 pKa = 4.3EE426 pKa = 4.41GDD428 pKa = 3.29VDD430 pKa = 5.22LSALSEE436 pKa = 4.54DD437 pKa = 5.33GEE439 pKa = 4.39DD440 pKa = 3.93QEE442 pKa = 5.51DD443 pKa = 3.67AAAEE447 pKa = 4.01AQEE450 pKa = 4.51SVTAAVEE457 pKa = 3.96TDD459 pKa = 3.62EE460 pKa = 4.78PVVDD464 pKa = 4.08VPLVNPLDD472 pKa = 5.1DD473 pKa = 6.05DD474 pKa = 5.02LNQPQVFLGG483 pKa = 3.63
Molecular weight: 50.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A171KPP8|A0A171KPP8_9BURK RimJ/RimL family protein N-acetyltransferase OS=Kerstersia gyiorum OX=206506 GN=AAV32_14080 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84AGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.89RR41 pKa = 11.84LAVV44 pKa = 3.5
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.99TRR25 pKa = 11.84AGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.89RR41 pKa = 11.84LAVV44 pKa = 3.5
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1083391 |
41 |
4541 |
331.6 |
36.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.465 ± 0.061 | 0.867 ± 0.013 |
5.28 ± 0.038 | 5.245 ± 0.038 |
3.444 ± 0.028 | 8.447 ± 0.048 |
2.182 ± 0.024 | 4.718 ± 0.03 |
2.715 ± 0.033 | 11.225 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.024 | 2.734 ± 0.031 |
5.113 ± 0.03 | 4.327 ± 0.03 |
7.002 ± 0.044 | 5.58 ± 0.031 |
4.926 ± 0.03 | 7.281 ± 0.045 |
1.499 ± 0.017 | 2.484 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
