
Eptesicus serotinus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyopsipapillomavirus; Dyopsipapillomavirus 1
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
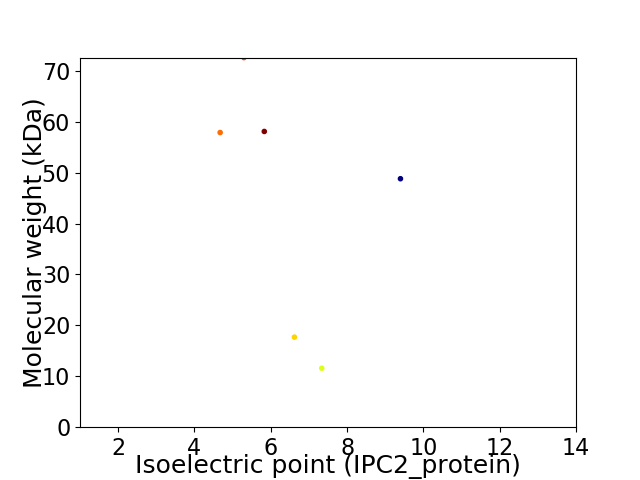
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|W8EH50|W8EH50_9PAPI Minor capsid protein L2 OS=Eptesicus serotinus papillomavirus 1 OX=1464071 GN=L2 PE=3 SV=1
MM1 pKa = 6.92VVVLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.69RR10 pKa = 11.84DD11 pKa = 3.34TVEE14 pKa = 3.89NIYY17 pKa = 9.95RR18 pKa = 11.84QCIRR22 pKa = 11.84FGTCPPDD29 pKa = 3.47VKK31 pKa = 11.2NKK33 pKa = 10.07VEE35 pKa = 4.06NNTLADD41 pKa = 4.04KK42 pKa = 10.55ILRR45 pKa = 11.84YY46 pKa = 10.17GSGAVYY52 pKa = 10.26FGGLGIGTGTGRR64 pKa = 11.84GTTGVSVGRR73 pKa = 11.84QVPVRR78 pKa = 11.84PFRR81 pKa = 11.84PHH83 pKa = 6.83GPIDD87 pKa = 3.61TIGIEE92 pKa = 4.26NIGPRR97 pKa = 11.84EE98 pKa = 4.08TPFAPPEE105 pKa = 4.02RR106 pKa = 11.84APVDD110 pKa = 3.33VDD112 pKa = 3.89LGTGHH117 pKa = 6.6VEE119 pKa = 3.83PSDD122 pKa = 3.59PSVIEE127 pKa = 4.09PTDD130 pKa = 3.86LPIDD134 pKa = 3.82IPTSSVDD141 pKa = 3.5EE142 pKa = 4.81VIVIPTPEE150 pKa = 3.67ITIGGDD156 pKa = 2.93TTAIIEE162 pKa = 4.38VTPSVPEE169 pKa = 3.61NTVISRR175 pKa = 11.84TQYY178 pKa = 9.63TNPAFDD184 pKa = 3.59ISLFSDD190 pKa = 3.52STSGEE195 pKa = 4.02LSASDD200 pKa = 4.76HH201 pKa = 5.87ITVSGGGGTIVGGHH215 pKa = 5.94YY216 pKa = 10.21PLAVPEE222 pKa = 4.48GEE224 pKa = 4.58EE225 pKa = 3.74IPLAEE230 pKa = 4.74FSPSVSRR237 pKa = 11.84FRR239 pKa = 11.84PGQVTEE245 pKa = 4.0QEE247 pKa = 4.15EE248 pKa = 4.77TSFITSTPEE257 pKa = 3.42GARR260 pKa = 11.84ARR262 pKa = 11.84PGRR265 pKa = 11.84TRR267 pKa = 11.84PSLYY271 pKa = 9.94GRR273 pKa = 11.84RR274 pKa = 11.84IEE276 pKa = 4.11QVEE279 pKa = 3.98ITDD282 pKa = 4.12PLFIDD287 pKa = 3.64EE288 pKa = 4.87PGRR291 pKa = 11.84LVEE294 pKa = 4.54FNYY297 pKa = 10.19VNPAFEE303 pKa = 4.63GDD305 pKa = 2.87ITEE308 pKa = 4.78IFEE311 pKa = 4.75QDD313 pKa = 3.03ILDD316 pKa = 3.98EE317 pKa = 4.41ARR319 pKa = 11.84AAPSSAFQDD328 pKa = 3.24IVRR331 pKa = 11.84LGRR334 pKa = 11.84QQLSRR339 pKa = 11.84SRR341 pKa = 11.84SGLVRR346 pKa = 11.84VSRR349 pKa = 11.84LGQRR353 pKa = 11.84GTIRR357 pKa = 11.84TRR359 pKa = 11.84SGVRR363 pKa = 11.84IGSTTHH369 pKa = 6.92FYY371 pKa = 10.55MDD373 pKa = 5.02LSPIGADD380 pKa = 2.85TGIPIEE386 pKa = 4.33PPGEE390 pKa = 3.98QSLEE394 pKa = 3.87ASIVQPFSEE403 pKa = 4.32EE404 pKa = 4.24GYY406 pKa = 11.0EE407 pKa = 4.3LVNLNDD413 pKa = 3.4VSGVVPDD420 pKa = 4.17EE421 pKa = 4.51SLLDD425 pKa = 3.78SYY427 pKa = 11.77EE428 pKa = 3.83EE429 pKa = 4.4TEE431 pKa = 5.28FIGSDD436 pKa = 3.65LQLVIHH442 pKa = 7.28DD443 pKa = 4.37EE444 pKa = 5.1DD445 pKa = 3.94IGIDD449 pKa = 3.61QDD451 pKa = 3.45SDD453 pKa = 3.95TVIDD457 pKa = 3.91IPSGLVKK464 pKa = 10.54KK465 pKa = 10.57AGPYY469 pKa = 10.17YY470 pKa = 9.98PDD472 pKa = 3.26IDD474 pKa = 3.86FAGVNIDD481 pKa = 3.35YY482 pKa = 10.4DD483 pKa = 3.69KK484 pKa = 11.45SHH486 pKa = 7.13TGRR489 pKa = 11.84AGDD492 pKa = 4.07IVPSDD497 pKa = 3.86TPIVTLDD504 pKa = 2.68IWSSDD509 pKa = 3.63FYY511 pKa = 11.06LHH513 pKa = 7.01PSLRR517 pKa = 11.84RR518 pKa = 11.84KK519 pKa = 9.13RR520 pKa = 11.84RR521 pKa = 11.84RR522 pKa = 11.84RR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 9.39YY526 pKa = 8.78VFVYY530 pKa = 10.68
MM1 pKa = 6.92VVVLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.69RR10 pKa = 11.84DD11 pKa = 3.34TVEE14 pKa = 3.89NIYY17 pKa = 9.95RR18 pKa = 11.84QCIRR22 pKa = 11.84FGTCPPDD29 pKa = 3.47VKK31 pKa = 11.2NKK33 pKa = 10.07VEE35 pKa = 4.06NNTLADD41 pKa = 4.04KK42 pKa = 10.55ILRR45 pKa = 11.84YY46 pKa = 10.17GSGAVYY52 pKa = 10.26FGGLGIGTGTGRR64 pKa = 11.84GTTGVSVGRR73 pKa = 11.84QVPVRR78 pKa = 11.84PFRR81 pKa = 11.84PHH83 pKa = 6.83GPIDD87 pKa = 3.61TIGIEE92 pKa = 4.26NIGPRR97 pKa = 11.84EE98 pKa = 4.08TPFAPPEE105 pKa = 4.02RR106 pKa = 11.84APVDD110 pKa = 3.33VDD112 pKa = 3.89LGTGHH117 pKa = 6.6VEE119 pKa = 3.83PSDD122 pKa = 3.59PSVIEE127 pKa = 4.09PTDD130 pKa = 3.86LPIDD134 pKa = 3.82IPTSSVDD141 pKa = 3.5EE142 pKa = 4.81VIVIPTPEE150 pKa = 3.67ITIGGDD156 pKa = 2.93TTAIIEE162 pKa = 4.38VTPSVPEE169 pKa = 3.61NTVISRR175 pKa = 11.84TQYY178 pKa = 9.63TNPAFDD184 pKa = 3.59ISLFSDD190 pKa = 3.52STSGEE195 pKa = 4.02LSASDD200 pKa = 4.76HH201 pKa = 5.87ITVSGGGGTIVGGHH215 pKa = 5.94YY216 pKa = 10.21PLAVPEE222 pKa = 4.48GEE224 pKa = 4.58EE225 pKa = 3.74IPLAEE230 pKa = 4.74FSPSVSRR237 pKa = 11.84FRR239 pKa = 11.84PGQVTEE245 pKa = 4.0QEE247 pKa = 4.15EE248 pKa = 4.77TSFITSTPEE257 pKa = 3.42GARR260 pKa = 11.84ARR262 pKa = 11.84PGRR265 pKa = 11.84TRR267 pKa = 11.84PSLYY271 pKa = 9.94GRR273 pKa = 11.84RR274 pKa = 11.84IEE276 pKa = 4.11QVEE279 pKa = 3.98ITDD282 pKa = 4.12PLFIDD287 pKa = 3.64EE288 pKa = 4.87PGRR291 pKa = 11.84LVEE294 pKa = 4.54FNYY297 pKa = 10.19VNPAFEE303 pKa = 4.63GDD305 pKa = 2.87ITEE308 pKa = 4.78IFEE311 pKa = 4.75QDD313 pKa = 3.03ILDD316 pKa = 3.98EE317 pKa = 4.41ARR319 pKa = 11.84AAPSSAFQDD328 pKa = 3.24IVRR331 pKa = 11.84LGRR334 pKa = 11.84QQLSRR339 pKa = 11.84SRR341 pKa = 11.84SGLVRR346 pKa = 11.84VSRR349 pKa = 11.84LGQRR353 pKa = 11.84GTIRR357 pKa = 11.84TRR359 pKa = 11.84SGVRR363 pKa = 11.84IGSTTHH369 pKa = 6.92FYY371 pKa = 10.55MDD373 pKa = 5.02LSPIGADD380 pKa = 2.85TGIPIEE386 pKa = 4.33PPGEE390 pKa = 3.98QSLEE394 pKa = 3.87ASIVQPFSEE403 pKa = 4.32EE404 pKa = 4.24GYY406 pKa = 11.0EE407 pKa = 4.3LVNLNDD413 pKa = 3.4VSGVVPDD420 pKa = 4.17EE421 pKa = 4.51SLLDD425 pKa = 3.78SYY427 pKa = 11.77EE428 pKa = 3.83EE429 pKa = 4.4TEE431 pKa = 5.28FIGSDD436 pKa = 3.65LQLVIHH442 pKa = 7.28DD443 pKa = 4.37EE444 pKa = 5.1DD445 pKa = 3.94IGIDD449 pKa = 3.61QDD451 pKa = 3.45SDD453 pKa = 3.95TVIDD457 pKa = 3.91IPSGLVKK464 pKa = 10.54KK465 pKa = 10.57AGPYY469 pKa = 10.17YY470 pKa = 9.98PDD472 pKa = 3.26IDD474 pKa = 3.86FAGVNIDD481 pKa = 3.35YY482 pKa = 10.4DD483 pKa = 3.69KK484 pKa = 11.45SHH486 pKa = 7.13TGRR489 pKa = 11.84AGDD492 pKa = 4.07IVPSDD497 pKa = 3.86TPIVTLDD504 pKa = 2.68IWSSDD509 pKa = 3.63FYY511 pKa = 11.06LHH513 pKa = 7.01PSLRR517 pKa = 11.84RR518 pKa = 11.84KK519 pKa = 9.13RR520 pKa = 11.84RR521 pKa = 11.84RR522 pKa = 11.84RR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 9.39YY526 pKa = 8.78VFVYY530 pKa = 10.68
Molecular weight: 57.89 kDa
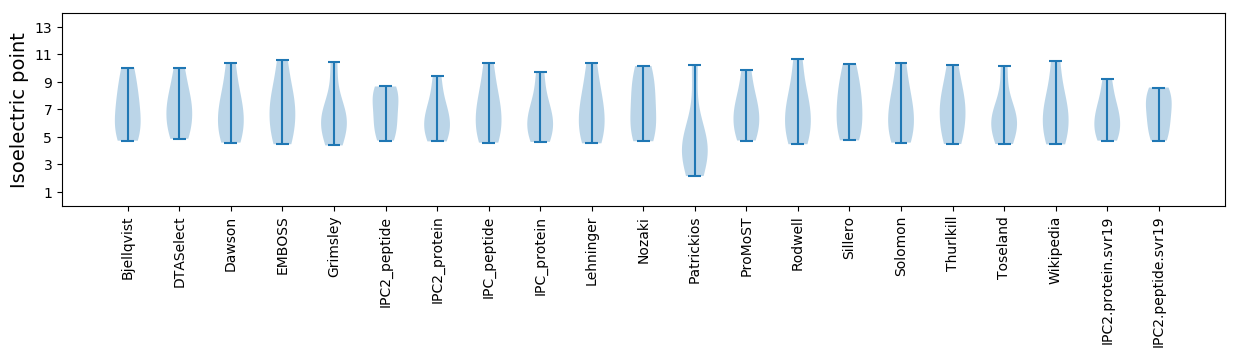
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8EAA5|W8EAA5_9PAPI Replication protein E1 OS=Eptesicus serotinus papillomavirus 1 OX=1464071 GN=E1 PE=3 SV=1
MM1 pKa = 7.82SEE3 pKa = 4.01LQSRR7 pKa = 11.84LDD9 pKa = 3.9SIQDD13 pKa = 3.6HH14 pKa = 6.06ILEE17 pKa = 4.34LFEE20 pKa = 5.12RR21 pKa = 11.84QSTDD25 pKa = 3.42LKK27 pKa = 11.17DD28 pKa = 3.78HH29 pKa = 5.5EE30 pKa = 4.91QYY32 pKa = 10.2WLLIRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.85QAILFLARR47 pKa = 11.84QKK49 pKa = 10.49HH50 pKa = 5.07VRR52 pKa = 11.84LSMPVPSQAASKK64 pKa = 10.62SRR66 pKa = 11.84ARR68 pKa = 11.84DD69 pKa = 3.74AIEE72 pKa = 3.68MSLLIKK78 pKa = 10.46GLSEE82 pKa = 4.01SQYY85 pKa = 9.96GTEE88 pKa = 4.18EE89 pKa = 3.63WTLQQVSRR97 pKa = 11.84EE98 pKa = 3.91RR99 pKa = 11.84LLAPPEE105 pKa = 3.92YY106 pKa = 9.49TFKK109 pKa = 11.02KK110 pKa = 8.9GGRR113 pKa = 11.84PITVMFDD120 pKa = 3.52DD121 pKa = 4.81MPDD124 pKa = 3.04NTAEE128 pKa = 3.95YY129 pKa = 8.52TAWDD133 pKa = 4.66LIYY136 pKa = 11.08YY137 pKa = 9.96QDD139 pKa = 6.11ADD141 pKa = 4.07NQWQKK146 pKa = 11.7AKK148 pKa = 10.81GDD150 pKa = 3.41VDD152 pKa = 3.91MEE154 pKa = 4.24GLFYY158 pKa = 10.73KK159 pKa = 10.55DD160 pKa = 3.43SEE162 pKa = 4.41GDD164 pKa = 2.98KK165 pKa = 10.81VYY167 pKa = 11.08YY168 pKa = 10.56VDD170 pKa = 5.19FSRR173 pKa = 11.84EE174 pKa = 3.68ATKK177 pKa = 10.75YY178 pKa = 10.31GRR180 pKa = 11.84TGKK183 pKa = 9.58WSLLYY188 pKa = 10.85NNNALASVDD197 pKa = 3.63SPGYY201 pKa = 9.64RR202 pKa = 11.84DD203 pKa = 3.87SDD205 pKa = 4.14TEE207 pKa = 4.18SSPDD211 pKa = 3.49SSPVPARR218 pKa = 11.84SSRR221 pKa = 11.84NSKK224 pKa = 10.11SRR226 pKa = 11.84SPAKK230 pKa = 9.37DD231 pKa = 2.85TRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84PRR237 pKa = 11.84LWLRR241 pKa = 11.84SKK243 pKa = 10.53SRR245 pKa = 11.84SRR247 pKa = 11.84SRR249 pKa = 11.84SRR251 pKa = 11.84SSSRR255 pKa = 11.84SSGRR259 pKa = 11.84SRR261 pKa = 11.84SRR263 pKa = 11.84SPFRR267 pKa = 11.84RR268 pKa = 11.84GASTVLTEE276 pKa = 4.06RR277 pKa = 11.84TRR279 pKa = 11.84RR280 pKa = 11.84SPGSHH285 pKa = 5.86LQSPGSPVSSTSAGRR300 pKa = 11.84RR301 pKa = 11.84GSAGPRR307 pKa = 11.84RR308 pKa = 11.84LGSSSRR314 pKa = 11.84PRR316 pKa = 11.84EE317 pKa = 3.85LAPDD321 pKa = 3.41QVGSSRR327 pKa = 11.84QTVSSRR333 pKa = 11.84PRR335 pKa = 11.84SSLEE339 pKa = 3.61RR340 pKa = 11.84LLEE343 pKa = 4.03EE344 pKa = 5.07ARR346 pKa = 11.84DD347 pKa = 3.95PPGLLLTGPPNTLKK361 pKa = 10.7CYY363 pKa = 10.22RR364 pKa = 11.84YY365 pKa = 9.78SLKK368 pKa = 10.61NKK370 pKa = 9.53HH371 pKa = 6.26SSLFTLISTTFHH383 pKa = 5.1WTEE386 pKa = 3.71SKK388 pKa = 10.44GPKK391 pKa = 9.7RR392 pKa = 11.84IGDD395 pKa = 3.57GKK397 pKa = 9.41MVIMFDD403 pKa = 4.46DD404 pKa = 3.93EE405 pKa = 4.58AARR408 pKa = 11.84EE409 pKa = 4.19WFLKK413 pKa = 9.54TVSLPRR419 pKa = 11.84TLAAVKK425 pKa = 10.5VDD427 pKa = 4.08FGGLL431 pKa = 3.32
MM1 pKa = 7.82SEE3 pKa = 4.01LQSRR7 pKa = 11.84LDD9 pKa = 3.9SIQDD13 pKa = 3.6HH14 pKa = 6.06ILEE17 pKa = 4.34LFEE20 pKa = 5.12RR21 pKa = 11.84QSTDD25 pKa = 3.42LKK27 pKa = 11.17DD28 pKa = 3.78HH29 pKa = 5.5EE30 pKa = 4.91QYY32 pKa = 10.2WLLIRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.85QAILFLARR47 pKa = 11.84QKK49 pKa = 10.49HH50 pKa = 5.07VRR52 pKa = 11.84LSMPVPSQAASKK64 pKa = 10.62SRR66 pKa = 11.84ARR68 pKa = 11.84DD69 pKa = 3.74AIEE72 pKa = 3.68MSLLIKK78 pKa = 10.46GLSEE82 pKa = 4.01SQYY85 pKa = 9.96GTEE88 pKa = 4.18EE89 pKa = 3.63WTLQQVSRR97 pKa = 11.84EE98 pKa = 3.91RR99 pKa = 11.84LLAPPEE105 pKa = 3.92YY106 pKa = 9.49TFKK109 pKa = 11.02KK110 pKa = 8.9GGRR113 pKa = 11.84PITVMFDD120 pKa = 3.52DD121 pKa = 4.81MPDD124 pKa = 3.04NTAEE128 pKa = 3.95YY129 pKa = 8.52TAWDD133 pKa = 4.66LIYY136 pKa = 11.08YY137 pKa = 9.96QDD139 pKa = 6.11ADD141 pKa = 4.07NQWQKK146 pKa = 11.7AKK148 pKa = 10.81GDD150 pKa = 3.41VDD152 pKa = 3.91MEE154 pKa = 4.24GLFYY158 pKa = 10.73KK159 pKa = 10.55DD160 pKa = 3.43SEE162 pKa = 4.41GDD164 pKa = 2.98KK165 pKa = 10.81VYY167 pKa = 11.08YY168 pKa = 10.56VDD170 pKa = 5.19FSRR173 pKa = 11.84EE174 pKa = 3.68ATKK177 pKa = 10.75YY178 pKa = 10.31GRR180 pKa = 11.84TGKK183 pKa = 9.58WSLLYY188 pKa = 10.85NNNALASVDD197 pKa = 3.63SPGYY201 pKa = 9.64RR202 pKa = 11.84DD203 pKa = 3.87SDD205 pKa = 4.14TEE207 pKa = 4.18SSPDD211 pKa = 3.49SSPVPARR218 pKa = 11.84SSRR221 pKa = 11.84NSKK224 pKa = 10.11SRR226 pKa = 11.84SPAKK230 pKa = 9.37DD231 pKa = 2.85TRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84PRR237 pKa = 11.84LWLRR241 pKa = 11.84SKK243 pKa = 10.53SRR245 pKa = 11.84SRR247 pKa = 11.84SRR249 pKa = 11.84SRR251 pKa = 11.84SSSRR255 pKa = 11.84SSGRR259 pKa = 11.84SRR261 pKa = 11.84SRR263 pKa = 11.84SPFRR267 pKa = 11.84RR268 pKa = 11.84GASTVLTEE276 pKa = 4.06RR277 pKa = 11.84TRR279 pKa = 11.84RR280 pKa = 11.84SPGSHH285 pKa = 5.86LQSPGSPVSSTSAGRR300 pKa = 11.84RR301 pKa = 11.84GSAGPRR307 pKa = 11.84RR308 pKa = 11.84LGSSSRR314 pKa = 11.84PRR316 pKa = 11.84EE317 pKa = 3.85LAPDD321 pKa = 3.41QVGSSRR327 pKa = 11.84QTVSSRR333 pKa = 11.84PRR335 pKa = 11.84SSLEE339 pKa = 3.61RR340 pKa = 11.84LLEE343 pKa = 4.03EE344 pKa = 5.07ARR346 pKa = 11.84DD347 pKa = 3.95PPGLLLTGPPNTLKK361 pKa = 10.7CYY363 pKa = 10.22RR364 pKa = 11.84YY365 pKa = 9.78SLKK368 pKa = 10.61NKK370 pKa = 9.53HH371 pKa = 6.26SSLFTLISTTFHH383 pKa = 5.1WTEE386 pKa = 3.71SKK388 pKa = 10.44GPKK391 pKa = 9.7RR392 pKa = 11.84IGDD395 pKa = 3.57GKK397 pKa = 9.41MVIMFDD403 pKa = 4.46DD404 pKa = 3.93EE405 pKa = 4.58AARR408 pKa = 11.84EE409 pKa = 4.19WFLKK413 pKa = 9.54TVSLPRR419 pKa = 11.84TLAAVKK425 pKa = 10.5VDD427 pKa = 4.08FGGLL431 pKa = 3.32
Molecular weight: 48.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2370 |
105 |
642 |
395.0 |
44.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.696 ± 0.58 | 1.646 ± 0.73 |
6.667 ± 0.338 | 6.287 ± 0.548 |
4.852 ± 0.568 | 6.54 ± 0.81 |
1.857 ± 0.211 | 4.979 ± 1.071 |
4.684 ± 0.959 | 8.776 ± 0.944 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.857 ± 0.38 | 3.713 ± 0.691 |
5.907 ± 1.01 | 4.262 ± 0.427 |
7.257 ± 1.072 | 8.692 ± 1.348 |
5.907 ± 0.43 | 6.371 ± 0.838 |
1.266 ± 0.333 | 2.785 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
