
Umatilla virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
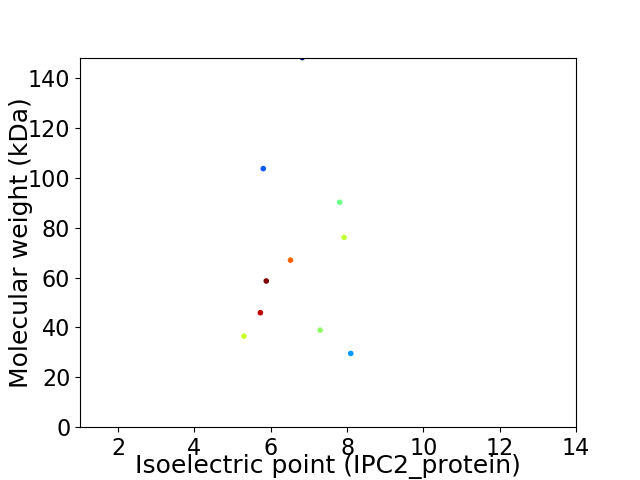
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8DP11|G8DP11_9REOV Non-structural protein NS3 OS=Umatilla virus OX=40060 PE=3 SV=1
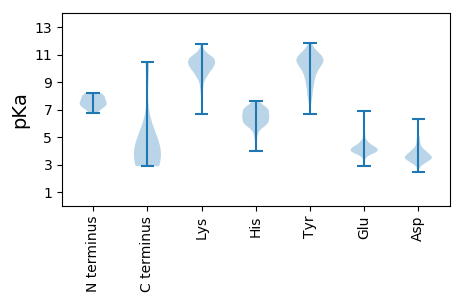
MM1 pKa = 7.88RR2 pKa = 11.84IVLLAPGDD10 pKa = 3.92IIEE13 pKa = 4.5KK14 pKa = 10.36SEE16 pKa = 4.29PEE18 pKa = 3.91LKK20 pKa = 10.29QRR22 pKa = 11.84GIDD25 pKa = 3.44IKK27 pKa = 11.0IKK29 pKa = 10.15DD30 pKa = 3.4WRR32 pKa = 11.84KK33 pKa = 10.4DD34 pKa = 3.39GGSHH38 pKa = 5.64VPSTEE43 pKa = 3.82RR44 pKa = 11.84EE45 pKa = 3.86NGGTGKK51 pKa = 10.12DD52 pKa = 3.79GEE54 pKa = 4.66SATPGSSGSNGHH66 pKa = 5.88VGRR69 pKa = 11.84DD70 pKa = 3.67GEE72 pKa = 4.27KK73 pKa = 10.48SKK75 pKa = 11.63GNGKK79 pKa = 9.42EE80 pKa = 4.25EE81 pKa = 4.12NDD83 pKa = 3.64SRR85 pKa = 11.84PGDD88 pKa = 3.9SRR90 pKa = 11.84CVRR93 pKa = 11.84TDD95 pKa = 3.1DD96 pKa = 3.73EE97 pKa = 5.9RR98 pKa = 11.84ITKK101 pKa = 8.76TEE103 pKa = 3.88IKK105 pKa = 10.68DD106 pKa = 3.47GDD108 pKa = 3.86GDD110 pKa = 3.92KK111 pKa = 11.51SGDD114 pKa = 3.7KK115 pKa = 10.55QSASAKK121 pKa = 9.7DD122 pKa = 3.57SKK124 pKa = 11.04DD125 pKa = 3.31AEE127 pKa = 4.34GDD129 pKa = 3.43EE130 pKa = 4.56RR131 pKa = 11.84SADD134 pKa = 3.58STVVKK139 pKa = 10.42GKK141 pKa = 10.22KK142 pKa = 9.07IKK144 pKa = 10.3EE145 pKa = 4.17NEE147 pKa = 4.05GTDD150 pKa = 3.62PKK152 pKa = 10.0ITKK155 pKa = 9.59QLAAEE160 pKa = 4.24RR161 pKa = 11.84SIDD164 pKa = 3.46GDD166 pKa = 3.99GPSGSVYY173 pKa = 10.63VLTRR177 pKa = 11.84AVADD181 pKa = 5.07AINQRR186 pKa = 11.84SGKK189 pKa = 9.42QLDD192 pKa = 3.78VLATLSDD199 pKa = 4.03LNGKK203 pKa = 7.65GKK205 pKa = 10.56CYY207 pKa = 10.13QISGGVGKK215 pKa = 10.44IIGLDD220 pKa = 3.08NDD222 pKa = 3.94AVKK225 pKa = 10.47EE226 pKa = 4.0QIDD229 pKa = 3.71ALNEE233 pKa = 3.95LKK235 pKa = 10.66SQYY238 pKa = 10.71DD239 pKa = 3.95GPNVNALLKK248 pKa = 10.02RR249 pKa = 11.84VLRR252 pKa = 11.84IEE254 pKa = 4.34SSSKK258 pKa = 9.97LDD260 pKa = 3.32QMFPKK265 pKa = 10.63NVDD268 pKa = 3.15EE269 pKa = 4.59KK270 pKa = 9.84QTIGSGVVLATNDD283 pKa = 3.71PSFLPQSNALFTCPTGDD300 pKa = 3.9TNWKK304 pKa = 9.14EE305 pKa = 4.27VARR308 pKa = 11.84QATNKK313 pKa = 9.25TSIRR317 pKa = 11.84AYY319 pKa = 8.31TYY321 pKa = 10.84SEE323 pKa = 3.97IGSGSEE329 pKa = 3.44IKK331 pKa = 10.43EE332 pKa = 4.17AFLSLIDD339 pKa = 3.84SLL341 pKa = 4.74
MM1 pKa = 7.88RR2 pKa = 11.84IVLLAPGDD10 pKa = 3.92IIEE13 pKa = 4.5KK14 pKa = 10.36SEE16 pKa = 4.29PEE18 pKa = 3.91LKK20 pKa = 10.29QRR22 pKa = 11.84GIDD25 pKa = 3.44IKK27 pKa = 11.0IKK29 pKa = 10.15DD30 pKa = 3.4WRR32 pKa = 11.84KK33 pKa = 10.4DD34 pKa = 3.39GGSHH38 pKa = 5.64VPSTEE43 pKa = 3.82RR44 pKa = 11.84EE45 pKa = 3.86NGGTGKK51 pKa = 10.12DD52 pKa = 3.79GEE54 pKa = 4.66SATPGSSGSNGHH66 pKa = 5.88VGRR69 pKa = 11.84DD70 pKa = 3.67GEE72 pKa = 4.27KK73 pKa = 10.48SKK75 pKa = 11.63GNGKK79 pKa = 9.42EE80 pKa = 4.25EE81 pKa = 4.12NDD83 pKa = 3.64SRR85 pKa = 11.84PGDD88 pKa = 3.9SRR90 pKa = 11.84CVRR93 pKa = 11.84TDD95 pKa = 3.1DD96 pKa = 3.73EE97 pKa = 5.9RR98 pKa = 11.84ITKK101 pKa = 8.76TEE103 pKa = 3.88IKK105 pKa = 10.68DD106 pKa = 3.47GDD108 pKa = 3.86GDD110 pKa = 3.92KK111 pKa = 11.51SGDD114 pKa = 3.7KK115 pKa = 10.55QSASAKK121 pKa = 9.7DD122 pKa = 3.57SKK124 pKa = 11.04DD125 pKa = 3.31AEE127 pKa = 4.34GDD129 pKa = 3.43EE130 pKa = 4.56RR131 pKa = 11.84SADD134 pKa = 3.58STVVKK139 pKa = 10.42GKK141 pKa = 10.22KK142 pKa = 9.07IKK144 pKa = 10.3EE145 pKa = 4.17NEE147 pKa = 4.05GTDD150 pKa = 3.62PKK152 pKa = 10.0ITKK155 pKa = 9.59QLAAEE160 pKa = 4.24RR161 pKa = 11.84SIDD164 pKa = 3.46GDD166 pKa = 3.99GPSGSVYY173 pKa = 10.63VLTRR177 pKa = 11.84AVADD181 pKa = 5.07AINQRR186 pKa = 11.84SGKK189 pKa = 9.42QLDD192 pKa = 3.78VLATLSDD199 pKa = 4.03LNGKK203 pKa = 7.65GKK205 pKa = 10.56CYY207 pKa = 10.13QISGGVGKK215 pKa = 10.44IIGLDD220 pKa = 3.08NDD222 pKa = 3.94AVKK225 pKa = 10.47EE226 pKa = 4.0QIDD229 pKa = 3.71ALNEE233 pKa = 3.95LKK235 pKa = 10.66SQYY238 pKa = 10.71DD239 pKa = 3.95GPNVNALLKK248 pKa = 10.02RR249 pKa = 11.84VLRR252 pKa = 11.84IEE254 pKa = 4.34SSSKK258 pKa = 9.97LDD260 pKa = 3.32QMFPKK265 pKa = 10.63NVDD268 pKa = 3.15EE269 pKa = 4.59KK270 pKa = 9.84QTIGSGVVLATNDD283 pKa = 3.71PSFLPQSNALFTCPTGDD300 pKa = 3.9TNWKK304 pKa = 9.14EE305 pKa = 4.27VARR308 pKa = 11.84QATNKK313 pKa = 9.25TSIRR317 pKa = 11.84AYY319 pKa = 8.31TYY321 pKa = 10.84SEE323 pKa = 3.97IGSGSEE329 pKa = 3.44IKK331 pKa = 10.43EE332 pKa = 4.17AFLSLIDD339 pKa = 3.84SLL341 pKa = 4.74
Molecular weight: 36.47 kDa
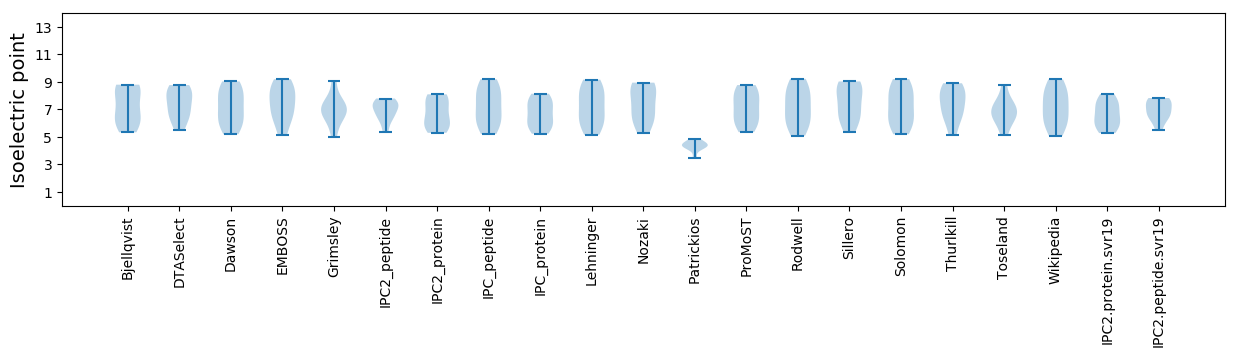
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8DP11|G8DP11_9REOV Non-structural protein NS3 OS=Umatilla virus OX=40060 PE=3 SV=1
MM1 pKa = 7.51LEE3 pKa = 3.85AAEE6 pKa = 4.22RR7 pKa = 11.84AQEE10 pKa = 3.86RR11 pKa = 11.84SKK13 pKa = 10.55ATEE16 pKa = 3.95SEE18 pKa = 4.47TVRR21 pKa = 11.84EE22 pKa = 4.19DD23 pKa = 3.31SDD25 pKa = 3.93NDD27 pKa = 3.83SMSKK31 pKa = 10.77GLLTEE36 pKa = 4.01TVLRR40 pKa = 11.84DD41 pKa = 3.62TKK43 pKa = 10.63RR44 pKa = 11.84GLGRR48 pKa = 11.84PVTQLTFVDD57 pKa = 4.52EE58 pKa = 4.44LSGGQGFMYY67 pKa = 10.3QQLQDD72 pKa = 3.31KK73 pKa = 10.05AQALNILTNAVTSTTGANDD92 pKa = 3.41TLKK95 pKa = 10.96NEE97 pKa = 4.17KK98 pKa = 10.21AVFGATSDD106 pKa = 3.76ALKK109 pKa = 10.5DD110 pKa = 3.87DD111 pKa = 4.1PFTRR115 pKa = 11.84YY116 pKa = 7.66TKK118 pKa = 10.14QLAYY122 pKa = 7.69EE123 pKa = 4.28TTISQILGQMATKK136 pKa = 9.57RR137 pKa = 11.84RR138 pKa = 11.84KK139 pKa = 8.83MLILDD144 pKa = 3.53VVKK147 pKa = 11.02YY148 pKa = 8.3MLGIILLMLLLWNMIVEE165 pKa = 4.71SIEE168 pKa = 3.85EE169 pKa = 3.88EE170 pKa = 4.38SYY172 pKa = 11.34KK173 pKa = 10.79EE174 pKa = 4.0VLSGMFGKK182 pKa = 10.31NNNDD186 pKa = 3.03TQALFSAARR195 pKa = 11.84AIRR198 pKa = 11.84RR199 pKa = 11.84YY200 pKa = 8.22TSYY203 pKa = 11.69LEE205 pKa = 4.37MISSIAMTFVVRR217 pKa = 11.84KK218 pKa = 10.06IMQLGVKK225 pKa = 10.04LKK227 pKa = 10.63ALEE230 pKa = 3.76RR231 pKa = 11.84DD232 pKa = 3.43KK233 pKa = 11.09IKK235 pKa = 10.89RR236 pKa = 11.84EE237 pKa = 4.07AYY239 pKa = 8.21NKK241 pKa = 8.14TVARR245 pKa = 11.84VSHH248 pKa = 6.46SGICSPSAPTLEE260 pKa = 4.27STEE263 pKa = 4.09RR264 pKa = 3.42
MM1 pKa = 7.51LEE3 pKa = 3.85AAEE6 pKa = 4.22RR7 pKa = 11.84AQEE10 pKa = 3.86RR11 pKa = 11.84SKK13 pKa = 10.55ATEE16 pKa = 3.95SEE18 pKa = 4.47TVRR21 pKa = 11.84EE22 pKa = 4.19DD23 pKa = 3.31SDD25 pKa = 3.93NDD27 pKa = 3.83SMSKK31 pKa = 10.77GLLTEE36 pKa = 4.01TVLRR40 pKa = 11.84DD41 pKa = 3.62TKK43 pKa = 10.63RR44 pKa = 11.84GLGRR48 pKa = 11.84PVTQLTFVDD57 pKa = 4.52EE58 pKa = 4.44LSGGQGFMYY67 pKa = 10.3QQLQDD72 pKa = 3.31KK73 pKa = 10.05AQALNILTNAVTSTTGANDD92 pKa = 3.41TLKK95 pKa = 10.96NEE97 pKa = 4.17KK98 pKa = 10.21AVFGATSDD106 pKa = 3.76ALKK109 pKa = 10.5DD110 pKa = 3.87DD111 pKa = 4.1PFTRR115 pKa = 11.84YY116 pKa = 7.66TKK118 pKa = 10.14QLAYY122 pKa = 7.69EE123 pKa = 4.28TTISQILGQMATKK136 pKa = 9.57RR137 pKa = 11.84RR138 pKa = 11.84KK139 pKa = 8.83MLILDD144 pKa = 3.53VVKK147 pKa = 11.02YY148 pKa = 8.3MLGIILLMLLLWNMIVEE165 pKa = 4.71SIEE168 pKa = 3.85EE169 pKa = 3.88EE170 pKa = 4.38SYY172 pKa = 11.34KK173 pKa = 10.79EE174 pKa = 4.0VLSGMFGKK182 pKa = 10.31NNNDD186 pKa = 3.03TQALFSAARR195 pKa = 11.84AIRR198 pKa = 11.84RR199 pKa = 11.84YY200 pKa = 8.22TSYY203 pKa = 11.69LEE205 pKa = 4.37MISSIAMTFVVRR217 pKa = 11.84KK218 pKa = 10.06IMQLGVKK225 pKa = 10.04LKK227 pKa = 10.63ALEE230 pKa = 3.76RR231 pKa = 11.84DD232 pKa = 3.43KK233 pKa = 11.09IKK235 pKa = 10.89RR236 pKa = 11.84EE237 pKa = 4.07AYY239 pKa = 8.21NKK241 pKa = 8.14TVARR245 pKa = 11.84VSHH248 pKa = 6.46SGICSPSAPTLEE260 pKa = 4.27STEE263 pKa = 4.09RR264 pKa = 3.42
Molecular weight: 29.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6098 |
264 |
1299 |
609.8 |
69.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.543 ± 0.391 | 1.23 ± 0.202 |
5.756 ± 0.35 | 6.756 ± 0.403 |
3.673 ± 0.376 | 5.805 ± 0.368 |
2.64 ± 0.271 | 6.642 ± 0.24 |
5.51 ± 0.675 | 9.347 ± 0.405 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.935 ± 0.207 | 4.51 ± 0.352 |
3.46 ± 0.433 | 4.182 ± 0.278 |
6.888 ± 0.345 | 6.953 ± 0.38 |
5.953 ± 0.323 | 6.264 ± 0.284 |
1.033 ± 0.178 | 3.919 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
