
Candidatus Vidania fulgoroideae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Vidania
Average proteome isoelectric point is 9.04
Get precalculated fractions of proteins
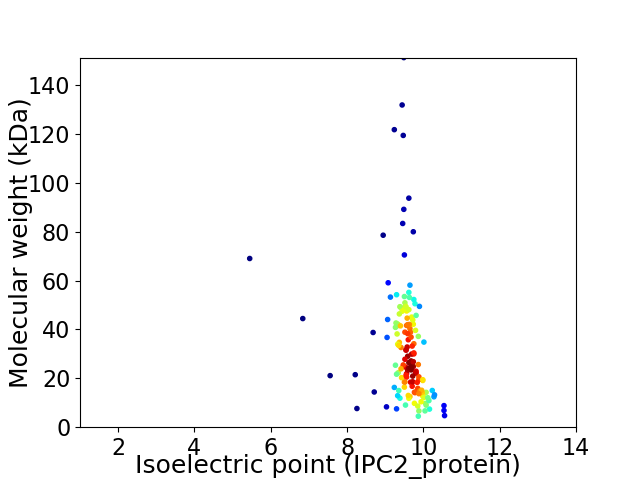
Virtual 2D-PAGE plot for 154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346E0L8|A0A346E0L8_9PROT tRNA-specific 2-thiouridylase MnmA OS=Candidatus Vidania fulgoroideae OX=881286 GN=C9I84_136 PE=4 SV=1
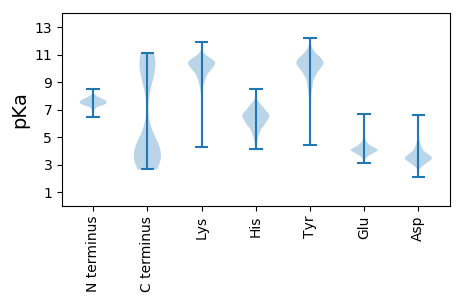
MM1 pKa = 6.82EE2 pKa = 4.81TSFGLYY8 pKa = 9.7IMNNIIGIDD17 pKa = 3.42LGTTNSCVSIMEE29 pKa = 4.39GNTAKK34 pKa = 10.74VIEE37 pKa = 4.28NSEE40 pKa = 4.47GYY42 pKa = 7.92RR43 pKa = 11.84TTPSVVAYY51 pKa = 10.55CNDD54 pKa = 3.91TILVGEE60 pKa = 4.25SAKK63 pKa = 10.53RR64 pKa = 11.84QSLTNPKK71 pKa = 8.55NTIFAVKK78 pKa = 10.02RR79 pKa = 11.84LIGRR83 pKa = 11.84KK84 pKa = 8.15FTDD87 pKa = 3.42PEE89 pKa = 3.79MLEE92 pKa = 4.28NIKK95 pKa = 9.68TSPYY99 pKa = 10.21KK100 pKa = 10.03IVKK103 pKa = 9.91AEE105 pKa = 4.4NGDD108 pKa = 3.13AWVEE112 pKa = 3.83IEE114 pKa = 4.28NGKK117 pKa = 10.25KK118 pKa = 9.42IAPQQVSSEE127 pKa = 4.06ILKK130 pKa = 10.61KK131 pKa = 9.62MKK133 pKa = 9.1KK134 pKa = 7.37TAEE137 pKa = 4.12SYY139 pKa = 10.92LGFEE143 pKa = 4.26IKK145 pKa = 10.25EE146 pKa = 4.06AVITVPAYY154 pKa = 10.64FNDD157 pKa = 3.76SQRR160 pKa = 11.84QATKK164 pKa = 10.34DD165 pKa = 2.98AGMIAGLKK173 pKa = 9.49INRR176 pKa = 11.84IVNEE180 pKa = 3.8PTAAALAFGLDD191 pKa = 3.5KK192 pKa = 11.27NIEE195 pKa = 4.09KK196 pKa = 10.2EE197 pKa = 4.16KK198 pKa = 10.86KK199 pKa = 9.32IVVYY203 pKa = 10.53DD204 pKa = 3.88LGGGTFDD211 pKa = 3.48ISVIEE216 pKa = 4.31ISMIDD221 pKa = 3.46NEE223 pKa = 4.42KK224 pKa = 10.37QFEE227 pKa = 4.31VLSTNGDD234 pKa = 3.7TLLGGEE240 pKa = 4.45DD241 pKa = 4.16FDD243 pKa = 5.27RR244 pKa = 11.84KK245 pKa = 9.96IIKK248 pKa = 9.84FLIDD252 pKa = 3.39KK253 pKa = 9.17YY254 pKa = 11.32LSIEE258 pKa = 4.57GIDD261 pKa = 3.7LEE263 pKa = 4.48KK264 pKa = 10.92DD265 pKa = 2.72ILALQRR271 pKa = 11.84LKK273 pKa = 10.74EE274 pKa = 4.03ASEE277 pKa = 4.1KK278 pKa = 10.74AKK280 pKa = 10.89KK281 pKa = 9.69EE282 pKa = 3.8LSNLEE287 pKa = 3.95STEE290 pKa = 3.65INLPYY295 pKa = 10.24ISINSSGPKK304 pKa = 9.42HH305 pKa = 5.85MKK307 pKa = 9.96ILLTRR312 pKa = 11.84AKK314 pKa = 10.74YY315 pKa = 10.07EE316 pKa = 3.83FLIEE320 pKa = 4.41DD321 pKa = 4.26LVKK324 pKa = 10.03KK325 pKa = 8.57TLIPCEE331 pKa = 3.96TALKK335 pKa = 10.49DD336 pKa = 3.61SLINISDD343 pKa = 3.45IDD345 pKa = 4.57DD346 pKa = 4.6IILVGGMTRR355 pKa = 11.84TPIIQKK361 pKa = 9.68RR362 pKa = 11.84VRR364 pKa = 11.84EE365 pKa = 4.04FFKK368 pKa = 11.01RR369 pKa = 11.84EE370 pKa = 3.63PKK372 pKa = 10.62KK373 pKa = 10.81NINPDD378 pKa = 3.03EE379 pKa = 4.18AVAIGASIQAQILSGDD395 pKa = 3.72RR396 pKa = 11.84KK397 pKa = 10.66DD398 pKa = 4.46VLLLDD403 pKa = 3.87VTPLSLGIEE412 pKa = 4.29TLGGVMTKK420 pKa = 9.89MIVKK424 pKa = 8.49NTTIPTKK431 pKa = 10.22CSQVFSTAEE440 pKa = 3.82EE441 pKa = 4.14NQNSVTIKK449 pKa = 9.68VYY451 pKa = 9.95QGEE454 pKa = 4.15RR455 pKa = 11.84SVAKK459 pKa = 9.57TNKK462 pKa = 10.23LLGEE466 pKa = 4.31FNLEE470 pKa = 4.23EE471 pKa = 4.48IEE473 pKa = 4.05PAPRR477 pKa = 11.84GVPQIEE483 pKa = 4.25VMFDD487 pKa = 2.82IDD489 pKa = 4.69SNGILNVSARR499 pKa = 11.84NKK501 pKa = 8.89VTGKK505 pKa = 9.64EE506 pKa = 3.77NKK508 pKa = 9.46IVIKK512 pKa = 10.67VNSGLTEE519 pKa = 3.9EE520 pKa = 5.2EE521 pKa = 3.92IEE523 pKa = 4.75NMINEE528 pKa = 4.17AKK530 pKa = 10.6KK531 pKa = 10.65NEE533 pKa = 4.14EE534 pKa = 3.66KK535 pKa = 10.99DD536 pKa = 3.62KK537 pKa = 10.64ITLEE541 pKa = 4.28TINLKK546 pKa = 10.62NKK548 pKa = 9.91IEE550 pKa = 3.86NLINQSEE557 pKa = 4.39KK558 pKa = 10.36IIKK561 pKa = 9.86EE562 pKa = 3.79NLNFIEE568 pKa = 4.37KK569 pKa = 10.12EE570 pKa = 4.01NYY572 pKa = 10.18LEE574 pKa = 5.62AEE576 pKa = 4.46TILNEE581 pKa = 4.37NKK583 pKa = 9.78ISYY586 pKa = 9.96LNKK589 pKa = 10.13KK590 pKa = 10.42KK591 pKa = 10.58EE592 pKa = 3.9DD593 pKa = 3.62LEE595 pKa = 4.56LNIINIEE602 pKa = 4.03NVMKK606 pKa = 10.64KK607 pKa = 10.7VNDD610 pKa = 4.54DD611 pKa = 2.99ILRR614 pKa = 11.84NKK616 pKa = 9.4KK617 pKa = 9.92
MM1 pKa = 6.82EE2 pKa = 4.81TSFGLYY8 pKa = 9.7IMNNIIGIDD17 pKa = 3.42LGTTNSCVSIMEE29 pKa = 4.39GNTAKK34 pKa = 10.74VIEE37 pKa = 4.28NSEE40 pKa = 4.47GYY42 pKa = 7.92RR43 pKa = 11.84TTPSVVAYY51 pKa = 10.55CNDD54 pKa = 3.91TILVGEE60 pKa = 4.25SAKK63 pKa = 10.53RR64 pKa = 11.84QSLTNPKK71 pKa = 8.55NTIFAVKK78 pKa = 10.02RR79 pKa = 11.84LIGRR83 pKa = 11.84KK84 pKa = 8.15FTDD87 pKa = 3.42PEE89 pKa = 3.79MLEE92 pKa = 4.28NIKK95 pKa = 9.68TSPYY99 pKa = 10.21KK100 pKa = 10.03IVKK103 pKa = 9.91AEE105 pKa = 4.4NGDD108 pKa = 3.13AWVEE112 pKa = 3.83IEE114 pKa = 4.28NGKK117 pKa = 10.25KK118 pKa = 9.42IAPQQVSSEE127 pKa = 4.06ILKK130 pKa = 10.61KK131 pKa = 9.62MKK133 pKa = 9.1KK134 pKa = 7.37TAEE137 pKa = 4.12SYY139 pKa = 10.92LGFEE143 pKa = 4.26IKK145 pKa = 10.25EE146 pKa = 4.06AVITVPAYY154 pKa = 10.64FNDD157 pKa = 3.76SQRR160 pKa = 11.84QATKK164 pKa = 10.34DD165 pKa = 2.98AGMIAGLKK173 pKa = 9.49INRR176 pKa = 11.84IVNEE180 pKa = 3.8PTAAALAFGLDD191 pKa = 3.5KK192 pKa = 11.27NIEE195 pKa = 4.09KK196 pKa = 10.2EE197 pKa = 4.16KK198 pKa = 10.86KK199 pKa = 9.32IVVYY203 pKa = 10.53DD204 pKa = 3.88LGGGTFDD211 pKa = 3.48ISVIEE216 pKa = 4.31ISMIDD221 pKa = 3.46NEE223 pKa = 4.42KK224 pKa = 10.37QFEE227 pKa = 4.31VLSTNGDD234 pKa = 3.7TLLGGEE240 pKa = 4.45DD241 pKa = 4.16FDD243 pKa = 5.27RR244 pKa = 11.84KK245 pKa = 9.96IIKK248 pKa = 9.84FLIDD252 pKa = 3.39KK253 pKa = 9.17YY254 pKa = 11.32LSIEE258 pKa = 4.57GIDD261 pKa = 3.7LEE263 pKa = 4.48KK264 pKa = 10.92DD265 pKa = 2.72ILALQRR271 pKa = 11.84LKK273 pKa = 10.74EE274 pKa = 4.03ASEE277 pKa = 4.1KK278 pKa = 10.74AKK280 pKa = 10.89KK281 pKa = 9.69EE282 pKa = 3.8LSNLEE287 pKa = 3.95STEE290 pKa = 3.65INLPYY295 pKa = 10.24ISINSSGPKK304 pKa = 9.42HH305 pKa = 5.85MKK307 pKa = 9.96ILLTRR312 pKa = 11.84AKK314 pKa = 10.74YY315 pKa = 10.07EE316 pKa = 3.83FLIEE320 pKa = 4.41DD321 pKa = 4.26LVKK324 pKa = 10.03KK325 pKa = 8.57TLIPCEE331 pKa = 3.96TALKK335 pKa = 10.49DD336 pKa = 3.61SLINISDD343 pKa = 3.45IDD345 pKa = 4.57DD346 pKa = 4.6IILVGGMTRR355 pKa = 11.84TPIIQKK361 pKa = 9.68RR362 pKa = 11.84VRR364 pKa = 11.84EE365 pKa = 4.04FFKK368 pKa = 11.01RR369 pKa = 11.84EE370 pKa = 3.63PKK372 pKa = 10.62KK373 pKa = 10.81NINPDD378 pKa = 3.03EE379 pKa = 4.18AVAIGASIQAQILSGDD395 pKa = 3.72RR396 pKa = 11.84KK397 pKa = 10.66DD398 pKa = 4.46VLLLDD403 pKa = 3.87VTPLSLGIEE412 pKa = 4.29TLGGVMTKK420 pKa = 9.89MIVKK424 pKa = 8.49NTTIPTKK431 pKa = 10.22CSQVFSTAEE440 pKa = 3.82EE441 pKa = 4.14NQNSVTIKK449 pKa = 9.68VYY451 pKa = 9.95QGEE454 pKa = 4.15RR455 pKa = 11.84SVAKK459 pKa = 9.57TNKK462 pKa = 10.23LLGEE466 pKa = 4.31FNLEE470 pKa = 4.23EE471 pKa = 4.48IEE473 pKa = 4.05PAPRR477 pKa = 11.84GVPQIEE483 pKa = 4.25VMFDD487 pKa = 2.82IDD489 pKa = 4.69SNGILNVSARR499 pKa = 11.84NKK501 pKa = 8.89VTGKK505 pKa = 9.64EE506 pKa = 3.77NKK508 pKa = 9.46IVIKK512 pKa = 10.67VNSGLTEE519 pKa = 3.9EE520 pKa = 5.2EE521 pKa = 3.92IEE523 pKa = 4.75NMINEE528 pKa = 4.17AKK530 pKa = 10.6KK531 pKa = 10.65NEE533 pKa = 4.14EE534 pKa = 3.66KK535 pKa = 10.99DD536 pKa = 3.62KK537 pKa = 10.64ITLEE541 pKa = 4.28TINLKK546 pKa = 10.62NKK548 pKa = 9.91IEE550 pKa = 3.86NLINQSEE557 pKa = 4.39KK558 pKa = 10.36IIKK561 pKa = 9.86EE562 pKa = 3.79NLNFIEE568 pKa = 4.37KK569 pKa = 10.12EE570 pKa = 4.01NYY572 pKa = 10.18LEE574 pKa = 5.62AEE576 pKa = 4.46TILNEE581 pKa = 4.37NKK583 pKa = 9.78ISYY586 pKa = 9.96LNKK589 pKa = 10.13KK590 pKa = 10.42KK591 pKa = 10.58EE592 pKa = 3.9DD593 pKa = 3.62LEE595 pKa = 4.56LNIINIEE602 pKa = 4.03NVMKK606 pKa = 10.64KK607 pKa = 10.7VNDD610 pKa = 4.54DD611 pKa = 2.99ILRR614 pKa = 11.84NKK616 pKa = 9.4KK617 pKa = 9.92
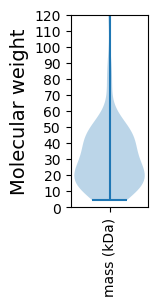
Molecular weight: 69.06 kDa
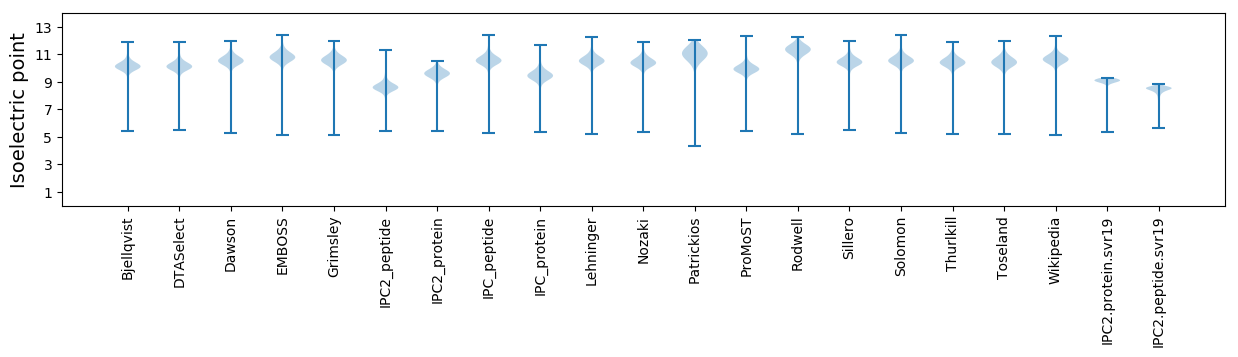
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346E0I9|A0A346E0I9_9PROT Uncharacterized protein OS=Candidatus Vidania fulgoroideae OX=881286 GN=C9I84_102 PE=4 SV=1
MM1 pKa = 7.21SRR3 pKa = 11.84VKK5 pKa = 10.61RR6 pKa = 11.84GVFSKK11 pKa = 10.51KK12 pKa = 7.65KK13 pKa = 8.23HH14 pKa = 5.34KK15 pKa = 10.55KK16 pKa = 8.88IIKK19 pKa = 8.27RR20 pKa = 11.84CKK22 pKa = 9.66GFTGRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.78NVFRR33 pKa = 11.84ISKK36 pKa = 8.49QAYY39 pKa = 8.26IKK41 pKa = 10.37SLQYY45 pKa = 10.12FYY47 pKa = 11.35RR48 pKa = 11.84DD49 pKa = 3.18TRR51 pKa = 11.84NKK53 pKa = 10.1KK54 pKa = 9.68RR55 pKa = 11.84DD56 pKa = 3.57FRR58 pKa = 11.84KK59 pKa = 9.99LWIININFYY68 pKa = 10.34LRR70 pKa = 11.84KK71 pKa = 10.15YY72 pKa = 8.44NIKK75 pKa = 8.76YY76 pKa = 10.45SRR78 pKa = 11.84FIKK81 pKa = 10.22YY82 pKa = 9.78IKK84 pKa = 9.71EE85 pKa = 3.65LNININRR92 pKa = 11.84KK93 pKa = 8.24SLYY96 pKa = 9.43YY97 pKa = 10.31LCKK100 pKa = 9.62EE101 pKa = 4.06YY102 pKa = 11.36NMKK105 pKa = 10.61KK106 pKa = 10.3LIIYY110 pKa = 10.06INEE113 pKa = 3.94KK114 pKa = 10.59KK115 pKa = 10.73YY116 pKa = 10.83LQNN119 pKa = 3.4
MM1 pKa = 7.21SRR3 pKa = 11.84VKK5 pKa = 10.61RR6 pKa = 11.84GVFSKK11 pKa = 10.51KK12 pKa = 7.65KK13 pKa = 8.23HH14 pKa = 5.34KK15 pKa = 10.55KK16 pKa = 8.88IIKK19 pKa = 8.27RR20 pKa = 11.84CKK22 pKa = 9.66GFTGRR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.78NVFRR33 pKa = 11.84ISKK36 pKa = 8.49QAYY39 pKa = 8.26IKK41 pKa = 10.37SLQYY45 pKa = 10.12FYY47 pKa = 11.35RR48 pKa = 11.84DD49 pKa = 3.18TRR51 pKa = 11.84NKK53 pKa = 10.1KK54 pKa = 9.68RR55 pKa = 11.84DD56 pKa = 3.57FRR58 pKa = 11.84KK59 pKa = 9.99LWIININFYY68 pKa = 10.34LRR70 pKa = 11.84KK71 pKa = 10.15YY72 pKa = 8.44NIKK75 pKa = 8.76YY76 pKa = 10.45SRR78 pKa = 11.84FIKK81 pKa = 10.22YY82 pKa = 9.78IKK84 pKa = 9.71EE85 pKa = 3.65LNININRR92 pKa = 11.84KK93 pKa = 8.24SLYY96 pKa = 9.43YY97 pKa = 10.31LCKK100 pKa = 9.62EE101 pKa = 4.06YY102 pKa = 11.36NMKK105 pKa = 10.61KK106 pKa = 10.3LIIYY110 pKa = 10.06INEE113 pKa = 3.94KK114 pKa = 10.59KK115 pKa = 10.73YY116 pKa = 10.83LQNN119 pKa = 3.4
Molecular weight: 15.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
42964 |
37 |
1292 |
279.0 |
33.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.543 ± 0.133 | 1.576 ± 0.055 |
2.795 ± 0.092 | 4.19 ± 0.178 |
7.595 ± 0.277 | 4.029 ± 0.184 |
0.968 ± 0.059 | 14.74 ± 0.187 |
19.156 ± 0.37 | 7.8 ± 0.126 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.536 ± 0.063 | 9.943 ± 0.189 |
1.769 ± 0.087 | 1.213 ± 0.063 |
2.9 ± 0.103 | 6.28 ± 0.156 |
3.182 ± 0.091 | 3.475 ± 0.125 |
0.314 ± 0.05 | 4.976 ± 0.131 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
