
Hubei tombus-like virus 26
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
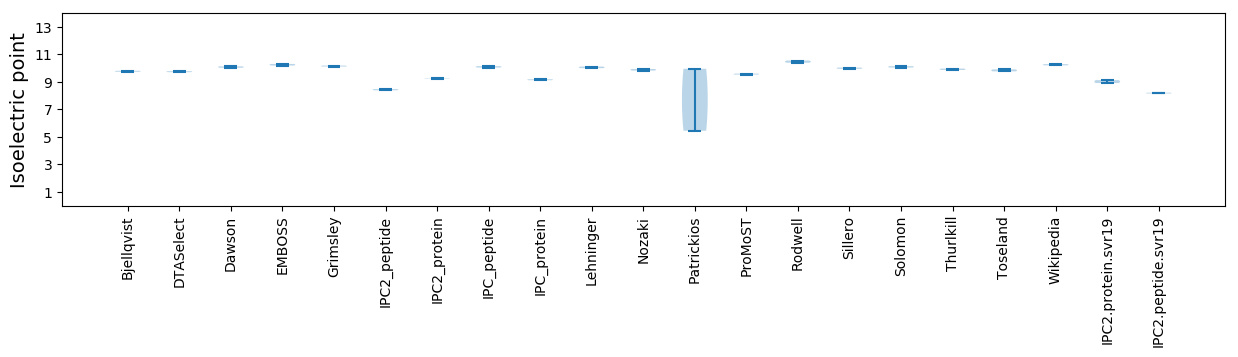
Average proteome isoelectric point is 9.02
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG47|A0A1L3KG47_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 26 OX=1923273 PE=4 SV=1
MM1 pKa = 7.24SQDD4 pKa = 3.69EE5 pKa = 4.91YY6 pKa = 11.0IKK8 pKa = 10.11TMPPKK13 pKa = 9.43YY14 pKa = 10.17SRR16 pKa = 11.84FLGVPIYY23 pKa = 10.59NVNNNVKK30 pKa = 10.36CFIKK34 pKa = 10.43KK35 pKa = 9.97EE36 pKa = 4.2KK37 pKa = 10.25ISLTKK42 pKa = 8.99YY43 pKa = 6.14TAPRR47 pKa = 11.84LIQHH51 pKa = 5.71NTMAYY56 pKa = 8.24NIEE59 pKa = 3.72VGRR62 pKa = 11.84YY63 pKa = 8.46IKK65 pKa = 10.33QLEE68 pKa = 4.1HH69 pKa = 7.28KK70 pKa = 9.54IYY72 pKa = 10.8QKK74 pKa = 11.04DD75 pKa = 3.58SSSQKK80 pKa = 10.68YY81 pKa = 7.4EE82 pKa = 3.79FCKK85 pKa = 10.37TDD87 pKa = 3.6LPEE90 pKa = 3.59MARR93 pKa = 11.84RR94 pKa = 11.84IMYY97 pKa = 9.13KK98 pKa = 10.02ASKK101 pKa = 8.91FQNPKK106 pKa = 9.86FVCIDD111 pKa = 3.57HH112 pKa = 6.43KK113 pKa = 11.1QFEE116 pKa = 4.27AHH118 pKa = 7.01LNGQHH123 pKa = 6.59INLEE127 pKa = 3.9NAFINWHH134 pKa = 5.39YY135 pKa = 10.6RR136 pKa = 11.84SPYY139 pKa = 9.27LQRR142 pKa = 11.84LLDD145 pKa = 3.53ARR147 pKa = 11.84TIFGTTTSGIKK158 pKa = 10.21YY159 pKa = 6.15RR160 pKa = 11.84TSMTRR165 pKa = 11.84TSGHH169 pKa = 5.62VKK171 pKa = 9.67TSFGNSILNLAFIKK185 pKa = 10.56HH186 pKa = 5.29VLKK189 pKa = 10.46QLKK192 pKa = 8.78IRR194 pKa = 11.84KK195 pKa = 8.95FEE197 pKa = 4.09VMVNGDD203 pKa = 3.56DD204 pKa = 4.36SIIIIEE210 pKa = 4.3NRR212 pKa = 11.84CHH214 pKa = 7.32LNEE217 pKa = 4.25SQLKK221 pKa = 10.07RR222 pKa = 11.84EE223 pKa = 3.89FRR225 pKa = 11.84KK226 pKa = 10.66LNMEE230 pKa = 4.19TKK232 pKa = 9.79IDD234 pKa = 3.71QITNNIYY241 pKa = 10.46DD242 pKa = 3.61VEE244 pKa = 4.33FCRR247 pKa = 11.84MKK249 pKa = 10.47ICIDD253 pKa = 3.54NAGKK257 pKa = 9.85PMMYY261 pKa = 9.08LTPDD265 pKa = 4.08RR266 pKa = 11.84IKK268 pKa = 10.84KK269 pKa = 9.93IFAQTHH275 pKa = 4.91KK276 pKa = 10.56PMRR279 pKa = 11.84RR280 pKa = 11.84NDD282 pKa = 3.52YY283 pKa = 10.86VRR285 pKa = 11.84DD286 pKa = 3.31LAYY289 pKa = 10.56ANSCIYY295 pKa = 10.77KK296 pKa = 10.23NLEE299 pKa = 3.9NYY301 pKa = 10.2HH302 pKa = 6.61SVFTDD307 pKa = 3.46IYY309 pKa = 9.94NHH311 pKa = 5.99YY312 pKa = 10.27NSLATTQLNKK322 pKa = 10.34YY323 pKa = 9.74LKK325 pKa = 10.47LEE327 pKa = 3.95PLILSLYY334 pKa = 9.46KK335 pKa = 10.44AASPDD340 pKa = 3.57PRR342 pKa = 11.84VSDD345 pKa = 3.72PMPHH349 pKa = 5.09YY350 pKa = 7.08TQPKK354 pKa = 8.25LSPPRR359 pKa = 11.84SMPNFKK365 pKa = 10.49LAVPKK370 pKa = 10.64SKK372 pKa = 10.73LSAGSQRR379 pKa = 11.84FLSRR383 pKa = 11.84MVSSSRR389 pKa = 11.84KK390 pKa = 9.19FIFPEE395 pKa = 4.17VFQEE399 pKa = 3.85TDD401 pKa = 2.56ATNITRR407 pKa = 11.84HH408 pKa = 5.28VYY410 pKa = 10.01INHH413 pKa = 6.02HH414 pKa = 5.26TQTLTVTHH422 pKa = 7.04GPRR425 pKa = 11.84PPEE428 pKa = 3.97PTSFPNPNLPFPTPMM443 pKa = 4.86
MM1 pKa = 7.24SQDD4 pKa = 3.69EE5 pKa = 4.91YY6 pKa = 11.0IKK8 pKa = 10.11TMPPKK13 pKa = 9.43YY14 pKa = 10.17SRR16 pKa = 11.84FLGVPIYY23 pKa = 10.59NVNNNVKK30 pKa = 10.36CFIKK34 pKa = 10.43KK35 pKa = 9.97EE36 pKa = 4.2KK37 pKa = 10.25ISLTKK42 pKa = 8.99YY43 pKa = 6.14TAPRR47 pKa = 11.84LIQHH51 pKa = 5.71NTMAYY56 pKa = 8.24NIEE59 pKa = 3.72VGRR62 pKa = 11.84YY63 pKa = 8.46IKK65 pKa = 10.33QLEE68 pKa = 4.1HH69 pKa = 7.28KK70 pKa = 9.54IYY72 pKa = 10.8QKK74 pKa = 11.04DD75 pKa = 3.58SSSQKK80 pKa = 10.68YY81 pKa = 7.4EE82 pKa = 3.79FCKK85 pKa = 10.37TDD87 pKa = 3.6LPEE90 pKa = 3.59MARR93 pKa = 11.84RR94 pKa = 11.84IMYY97 pKa = 9.13KK98 pKa = 10.02ASKK101 pKa = 8.91FQNPKK106 pKa = 9.86FVCIDD111 pKa = 3.57HH112 pKa = 6.43KK113 pKa = 11.1QFEE116 pKa = 4.27AHH118 pKa = 7.01LNGQHH123 pKa = 6.59INLEE127 pKa = 3.9NAFINWHH134 pKa = 5.39YY135 pKa = 10.6RR136 pKa = 11.84SPYY139 pKa = 9.27LQRR142 pKa = 11.84LLDD145 pKa = 3.53ARR147 pKa = 11.84TIFGTTTSGIKK158 pKa = 10.21YY159 pKa = 6.15RR160 pKa = 11.84TSMTRR165 pKa = 11.84TSGHH169 pKa = 5.62VKK171 pKa = 9.67TSFGNSILNLAFIKK185 pKa = 10.56HH186 pKa = 5.29VLKK189 pKa = 10.46QLKK192 pKa = 8.78IRR194 pKa = 11.84KK195 pKa = 8.95FEE197 pKa = 4.09VMVNGDD203 pKa = 3.56DD204 pKa = 4.36SIIIIEE210 pKa = 4.3NRR212 pKa = 11.84CHH214 pKa = 7.32LNEE217 pKa = 4.25SQLKK221 pKa = 10.07RR222 pKa = 11.84EE223 pKa = 3.89FRR225 pKa = 11.84KK226 pKa = 10.66LNMEE230 pKa = 4.19TKK232 pKa = 9.79IDD234 pKa = 3.71QITNNIYY241 pKa = 10.46DD242 pKa = 3.61VEE244 pKa = 4.33FCRR247 pKa = 11.84MKK249 pKa = 10.47ICIDD253 pKa = 3.54NAGKK257 pKa = 9.85PMMYY261 pKa = 9.08LTPDD265 pKa = 4.08RR266 pKa = 11.84IKK268 pKa = 10.84KK269 pKa = 9.93IFAQTHH275 pKa = 4.91KK276 pKa = 10.56PMRR279 pKa = 11.84RR280 pKa = 11.84NDD282 pKa = 3.52YY283 pKa = 10.86VRR285 pKa = 11.84DD286 pKa = 3.31LAYY289 pKa = 10.56ANSCIYY295 pKa = 10.77KK296 pKa = 10.23NLEE299 pKa = 3.9NYY301 pKa = 10.2HH302 pKa = 6.61SVFTDD307 pKa = 3.46IYY309 pKa = 9.94NHH311 pKa = 5.99YY312 pKa = 10.27NSLATTQLNKK322 pKa = 10.34YY323 pKa = 9.74LKK325 pKa = 10.47LEE327 pKa = 3.95PLILSLYY334 pKa = 9.46KK335 pKa = 10.44AASPDD340 pKa = 3.57PRR342 pKa = 11.84VSDD345 pKa = 3.72PMPHH349 pKa = 5.09YY350 pKa = 7.08TQPKK354 pKa = 8.25LSPPRR359 pKa = 11.84SMPNFKK365 pKa = 10.49LAVPKK370 pKa = 10.64SKK372 pKa = 10.73LSAGSQRR379 pKa = 11.84FLSRR383 pKa = 11.84MVSSSRR389 pKa = 11.84KK390 pKa = 9.19FIFPEE395 pKa = 4.17VFQEE399 pKa = 3.85TDD401 pKa = 2.56ATNITRR407 pKa = 11.84HH408 pKa = 5.28VYY410 pKa = 10.01INHH413 pKa = 6.02HH414 pKa = 5.26TQTLTVTHH422 pKa = 7.04GPRR425 pKa = 11.84PPEE428 pKa = 3.97PTSFPNPNLPFPTPMM443 pKa = 4.86
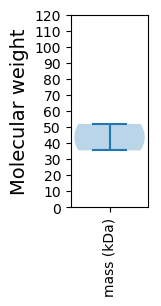
Molecular weight: 51.86 kDa
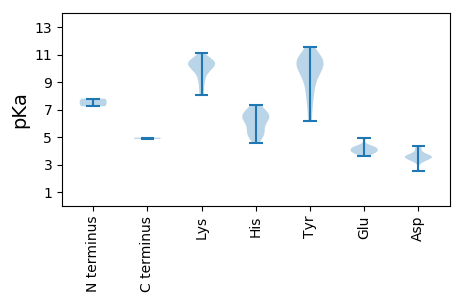
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG47|A0A1L3KG47_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 26 OX=1923273 PE=4 SV=1
MM1 pKa = 7.79ALPKK5 pKa = 10.44ASEE8 pKa = 4.21TLSPQDD14 pKa = 3.75FPNPPKK20 pKa = 9.88TLAPIPSLPDD30 pKa = 3.48SLPKK34 pKa = 10.2SLKK37 pKa = 9.97HH38 pKa = 6.37SRR40 pKa = 11.84KK41 pKa = 8.27CTPGTFDD48 pKa = 3.31HH49 pKa = 6.49QRR51 pKa = 11.84LASEE55 pKa = 4.43LEE57 pKa = 4.23GVVTCNNTYY66 pKa = 8.1THH68 pKa = 6.74LHH70 pKa = 5.85DD71 pKa = 4.12TVNIHH76 pKa = 6.7TNLQDD81 pKa = 3.43EE82 pKa = 4.9LNAHH86 pKa = 6.88LEE88 pKa = 3.93LAMALKK94 pKa = 9.77PRR96 pKa = 11.84NVDD99 pKa = 4.23FYY101 pKa = 10.5RR102 pKa = 11.84QAIGEE107 pKa = 4.32SQNFLNRR114 pKa = 11.84YY115 pKa = 7.85NVNLPSARR123 pKa = 11.84LLLIRR128 pKa = 11.84NSLDD132 pKa = 3.26FLSSLTQTEE141 pKa = 4.0QLIHH145 pKa = 6.46SKK147 pKa = 9.71VEE149 pKa = 3.83KK150 pKa = 8.04TQKK153 pKa = 10.38FSGLKK158 pKa = 9.38YY159 pKa = 10.0IKK161 pKa = 10.04EE162 pKa = 4.07NSLSKK167 pKa = 10.85VNHH170 pKa = 6.35ILSKK174 pKa = 10.17IASRR178 pKa = 11.84PDD180 pKa = 3.23FRR182 pKa = 11.84LKK184 pKa = 10.56AHH186 pKa = 6.25VSRR189 pKa = 11.84SYY191 pKa = 11.56KK192 pKa = 10.56LYY194 pKa = 9.18TQLKK198 pKa = 8.3SHH200 pKa = 6.37IHH202 pKa = 4.6YY203 pKa = 10.45QYY205 pKa = 10.33TLTLIPITLIFSILTLPLTILRR227 pKa = 11.84ALQTRR232 pKa = 11.84WRR234 pKa = 11.84ASSTGTTTTVFLNPMFSYY252 pKa = 10.77VLPKK256 pKa = 10.36FSATSQNKK264 pKa = 8.35PLKK267 pKa = 10.42ASEE270 pKa = 3.67IYY272 pKa = 10.19LRR274 pKa = 11.84HH275 pKa = 4.97TAYY278 pKa = 10.56AEE280 pKa = 4.0TSAPCRR286 pKa = 11.84KK287 pKa = 9.14MNILKK292 pKa = 10.34LCLLSTLAFLVFLSIMLITMM312 pKa = 4.96
MM1 pKa = 7.79ALPKK5 pKa = 10.44ASEE8 pKa = 4.21TLSPQDD14 pKa = 3.75FPNPPKK20 pKa = 9.88TLAPIPSLPDD30 pKa = 3.48SLPKK34 pKa = 10.2SLKK37 pKa = 9.97HH38 pKa = 6.37SRR40 pKa = 11.84KK41 pKa = 8.27CTPGTFDD48 pKa = 3.31HH49 pKa = 6.49QRR51 pKa = 11.84LASEE55 pKa = 4.43LEE57 pKa = 4.23GVVTCNNTYY66 pKa = 8.1THH68 pKa = 6.74LHH70 pKa = 5.85DD71 pKa = 4.12TVNIHH76 pKa = 6.7TNLQDD81 pKa = 3.43EE82 pKa = 4.9LNAHH86 pKa = 6.88LEE88 pKa = 3.93LAMALKK94 pKa = 9.77PRR96 pKa = 11.84NVDD99 pKa = 4.23FYY101 pKa = 10.5RR102 pKa = 11.84QAIGEE107 pKa = 4.32SQNFLNRR114 pKa = 11.84YY115 pKa = 7.85NVNLPSARR123 pKa = 11.84LLLIRR128 pKa = 11.84NSLDD132 pKa = 3.26FLSSLTQTEE141 pKa = 4.0QLIHH145 pKa = 6.46SKK147 pKa = 9.71VEE149 pKa = 3.83KK150 pKa = 8.04TQKK153 pKa = 10.38FSGLKK158 pKa = 9.38YY159 pKa = 10.0IKK161 pKa = 10.04EE162 pKa = 4.07NSLSKK167 pKa = 10.85VNHH170 pKa = 6.35ILSKK174 pKa = 10.17IASRR178 pKa = 11.84PDD180 pKa = 3.23FRR182 pKa = 11.84LKK184 pKa = 10.56AHH186 pKa = 6.25VSRR189 pKa = 11.84SYY191 pKa = 11.56KK192 pKa = 10.56LYY194 pKa = 9.18TQLKK198 pKa = 8.3SHH200 pKa = 6.37IHH202 pKa = 4.6YY203 pKa = 10.45QYY205 pKa = 10.33TLTLIPITLIFSILTLPLTILRR227 pKa = 11.84ALQTRR232 pKa = 11.84WRR234 pKa = 11.84ASSTGTTTTVFLNPMFSYY252 pKa = 10.77VLPKK256 pKa = 10.36FSATSQNKK264 pKa = 8.35PLKK267 pKa = 10.42ASEE270 pKa = 3.67IYY272 pKa = 10.19LRR274 pKa = 11.84HH275 pKa = 4.97TAYY278 pKa = 10.56AEE280 pKa = 4.0TSAPCRR286 pKa = 11.84KK287 pKa = 9.14MNILKK292 pKa = 10.34LCLLSTLAFLVFLSIMLITMM312 pKa = 4.96
Molecular weight: 35.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
755 |
312 |
443 |
377.5 |
43.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.901 ± 0.717 | 1.457 ± 0.105 |
3.311 ± 0.451 | 3.974 ± 0.27 |
4.636 ± 0.283 | 2.119 ± 0.312 |
3.841 ± 0.003 | 6.887 ± 0.674 |
7.947 ± 0.733 | 10.861 ± 2.921 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.914 ± 0.597 | 6.49 ± 0.628 |
6.49 ± 0.241 | 3.974 ± 0.077 |
5.43 ± 0.376 | 8.212 ± 1.04 |
7.815 ± 0.893 | 3.841 ± 0.19 |
0.265 ± 0.034 | 4.636 ± 0.669 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
