
Anaerobiospirillum thomasii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Aeromonadales; Succinivibrionaceae; Anaerobiospirillum
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
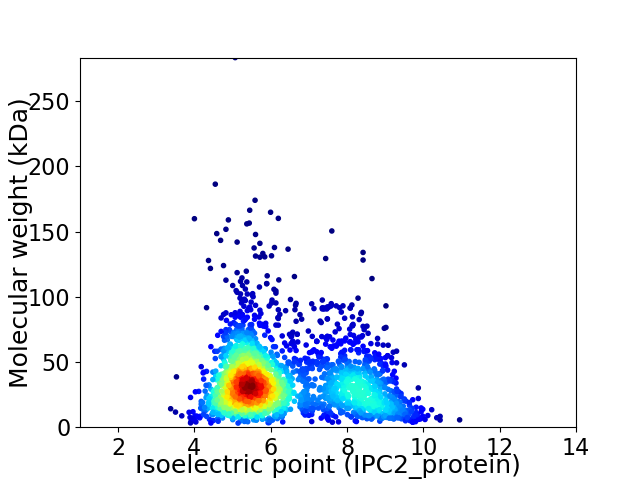
Virtual 2D-PAGE plot for 2368 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2X0VA45|A0A2X0VA45_9GAMM Flagellin OS=Anaerobiospirillum thomasii OX=179995 GN=flaD_7 PE=3 SV=1
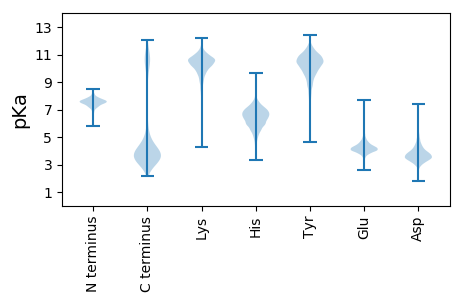
MM1 pKa = 7.66SYY3 pKa = 11.02INIVLDD9 pKa = 4.44GYY11 pKa = 12.01VMDD14 pKa = 5.82LNMDD18 pKa = 3.81KK19 pKa = 10.49LTDD22 pKa = 3.67EE23 pKa = 4.3QVLEE27 pKa = 4.46FKK29 pKa = 10.9DD30 pKa = 3.53AVRR33 pKa = 11.84AYY35 pKa = 10.71DD36 pKa = 3.96KK37 pKa = 11.41DD38 pKa = 3.36PDD40 pKa = 3.67ANYY43 pKa = 10.14IEE45 pKa = 5.34DD46 pKa = 4.69LDD48 pKa = 4.02IFYY51 pKa = 7.92DD52 pKa = 3.77TQINADD58 pKa = 3.8TVQTLRR64 pKa = 11.84GYY66 pKa = 11.57YY67 pKa = 10.39DD68 pKa = 3.76FALDD72 pKa = 3.35AYY74 pKa = 10.65LRR76 pKa = 11.84RR77 pKa = 11.84FDD79 pKa = 6.14DD80 pKa = 4.33DD81 pKa = 3.74GQRR84 pKa = 11.84ITDD87 pKa = 3.96LEE89 pKa = 5.55GDD91 pKa = 3.67DD92 pKa = 3.82EE93 pKa = 5.18VFFEE97 pKa = 5.33EE98 pKa = 4.73IASSAADD105 pKa = 3.39DD106 pKa = 3.97PSFLEE111 pKa = 4.94EE112 pKa = 4.32ISGLDD117 pKa = 4.38LEE119 pKa = 4.86DD120 pKa = 3.88GYY122 pKa = 11.75YY123 pKa = 10.18FVAQRR128 pKa = 11.84VSKK131 pKa = 9.69CTDD134 pKa = 2.94SFIVEE139 pKa = 4.28CDD141 pKa = 3.42EE142 pKa = 5.88EE143 pKa = 5.22IDD145 pKa = 3.95EE146 pKa = 5.19LDD148 pKa = 3.71PFALEE153 pKa = 4.48DD154 pKa = 3.56EE155 pKa = 4.69VAIDD159 pKa = 3.43VCTISEE165 pKa = 4.66LPTVMGRR172 pKa = 11.84EE173 pKa = 4.24INTDD177 pKa = 2.9SYY179 pKa = 11.99NIITNMSYY187 pKa = 10.28KK188 pKa = 10.69GEE190 pKa = 4.12YY191 pKa = 9.49FLNEE195 pKa = 3.7VDD197 pKa = 4.28EE198 pKa = 4.61SPIYY202 pKa = 9.57DD203 pKa = 4.07TIFSTDD209 pKa = 3.03KK210 pKa = 10.9VKK212 pKa = 11.05EE213 pKa = 3.88DD214 pKa = 3.38VLIIFQVVEE223 pKa = 3.92GHH225 pKa = 6.67LSIVYY230 pKa = 9.08TYY232 pKa = 11.12DD233 pKa = 3.21AFSDD237 pKa = 4.2EE238 pKa = 4.69PEE240 pKa = 4.21HH241 pKa = 7.4FYY243 pKa = 11.83DD244 pKa = 5.92HH245 pKa = 7.15IDD247 pKa = 3.37KK248 pKa = 9.82TLVDD252 pKa = 3.61MPIYY256 pKa = 10.19QNNLIQQIYY265 pKa = 10.59LEE267 pKa = 4.54GDD269 pKa = 3.23DD270 pKa = 5.67YY271 pKa = 11.86YY272 pKa = 11.51DD273 pKa = 3.76EE274 pKa = 6.46DD275 pKa = 4.1EE276 pKa = 5.63VDD278 pKa = 5.24DD279 pKa = 4.85EE280 pKa = 6.33DD281 pKa = 6.35YY282 pKa = 11.62DD283 pKa = 5.84DD284 pKa = 5.29EE285 pKa = 6.99DD286 pKa = 4.81SFDD289 pKa = 5.06GEE291 pKa = 5.15DD292 pKa = 3.78FDD294 pKa = 6.15EE295 pKa = 5.66SLDD298 pKa = 4.89DD299 pKa = 6.14DD300 pKa = 5.26YY301 pKa = 12.06EE302 pKa = 6.59DD303 pKa = 6.32DD304 pKa = 4.25DD305 pKa = 6.06DD306 pKa = 5.43EE307 pKa = 4.63YY308 pKa = 11.84ASSRR312 pKa = 11.84KK313 pKa = 9.16KK314 pKa = 10.45DD315 pKa = 3.11RR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84YY319 pKa = 9.86EE320 pKa = 3.86GGKK323 pKa = 9.53SDD325 pKa = 5.52KK326 pKa = 10.92EE327 pKa = 3.92DD328 pKa = 4.07DD329 pKa = 3.81NFF331 pKa = 4.78
MM1 pKa = 7.66SYY3 pKa = 11.02INIVLDD9 pKa = 4.44GYY11 pKa = 12.01VMDD14 pKa = 5.82LNMDD18 pKa = 3.81KK19 pKa = 10.49LTDD22 pKa = 3.67EE23 pKa = 4.3QVLEE27 pKa = 4.46FKK29 pKa = 10.9DD30 pKa = 3.53AVRR33 pKa = 11.84AYY35 pKa = 10.71DD36 pKa = 3.96KK37 pKa = 11.41DD38 pKa = 3.36PDD40 pKa = 3.67ANYY43 pKa = 10.14IEE45 pKa = 5.34DD46 pKa = 4.69LDD48 pKa = 4.02IFYY51 pKa = 7.92DD52 pKa = 3.77TQINADD58 pKa = 3.8TVQTLRR64 pKa = 11.84GYY66 pKa = 11.57YY67 pKa = 10.39DD68 pKa = 3.76FALDD72 pKa = 3.35AYY74 pKa = 10.65LRR76 pKa = 11.84RR77 pKa = 11.84FDD79 pKa = 6.14DD80 pKa = 4.33DD81 pKa = 3.74GQRR84 pKa = 11.84ITDD87 pKa = 3.96LEE89 pKa = 5.55GDD91 pKa = 3.67DD92 pKa = 3.82EE93 pKa = 5.18VFFEE97 pKa = 5.33EE98 pKa = 4.73IASSAADD105 pKa = 3.39DD106 pKa = 3.97PSFLEE111 pKa = 4.94EE112 pKa = 4.32ISGLDD117 pKa = 4.38LEE119 pKa = 4.86DD120 pKa = 3.88GYY122 pKa = 11.75YY123 pKa = 10.18FVAQRR128 pKa = 11.84VSKK131 pKa = 9.69CTDD134 pKa = 2.94SFIVEE139 pKa = 4.28CDD141 pKa = 3.42EE142 pKa = 5.88EE143 pKa = 5.22IDD145 pKa = 3.95EE146 pKa = 5.19LDD148 pKa = 3.71PFALEE153 pKa = 4.48DD154 pKa = 3.56EE155 pKa = 4.69VAIDD159 pKa = 3.43VCTISEE165 pKa = 4.66LPTVMGRR172 pKa = 11.84EE173 pKa = 4.24INTDD177 pKa = 2.9SYY179 pKa = 11.99NIITNMSYY187 pKa = 10.28KK188 pKa = 10.69GEE190 pKa = 4.12YY191 pKa = 9.49FLNEE195 pKa = 3.7VDD197 pKa = 4.28EE198 pKa = 4.61SPIYY202 pKa = 9.57DD203 pKa = 4.07TIFSTDD209 pKa = 3.03KK210 pKa = 10.9VKK212 pKa = 11.05EE213 pKa = 3.88DD214 pKa = 3.38VLIIFQVVEE223 pKa = 3.92GHH225 pKa = 6.67LSIVYY230 pKa = 9.08TYY232 pKa = 11.12DD233 pKa = 3.21AFSDD237 pKa = 4.2EE238 pKa = 4.69PEE240 pKa = 4.21HH241 pKa = 7.4FYY243 pKa = 11.83DD244 pKa = 5.92HH245 pKa = 7.15IDD247 pKa = 3.37KK248 pKa = 9.82TLVDD252 pKa = 3.61MPIYY256 pKa = 10.19QNNLIQQIYY265 pKa = 10.59LEE267 pKa = 4.54GDD269 pKa = 3.23DD270 pKa = 5.67YY271 pKa = 11.86YY272 pKa = 11.51DD273 pKa = 3.76EE274 pKa = 6.46DD275 pKa = 4.1EE276 pKa = 5.63VDD278 pKa = 5.24DD279 pKa = 4.85EE280 pKa = 6.33DD281 pKa = 6.35YY282 pKa = 11.62DD283 pKa = 5.84DD284 pKa = 5.29EE285 pKa = 6.99DD286 pKa = 4.81SFDD289 pKa = 5.06GEE291 pKa = 5.15DD292 pKa = 3.78FDD294 pKa = 6.15EE295 pKa = 5.66SLDD298 pKa = 4.89DD299 pKa = 6.14DD300 pKa = 5.26YY301 pKa = 12.06EE302 pKa = 6.59DD303 pKa = 6.32DD304 pKa = 4.25DD305 pKa = 6.06DD306 pKa = 5.43EE307 pKa = 4.63YY308 pKa = 11.84ASSRR312 pKa = 11.84KK313 pKa = 9.16KK314 pKa = 10.45DD315 pKa = 3.11RR316 pKa = 11.84RR317 pKa = 11.84RR318 pKa = 11.84YY319 pKa = 9.86EE320 pKa = 3.86GGKK323 pKa = 9.53SDD325 pKa = 5.52KK326 pKa = 10.92EE327 pKa = 3.92DD328 pKa = 4.07DD329 pKa = 3.81NFF331 pKa = 4.78
Molecular weight: 38.61 kDa
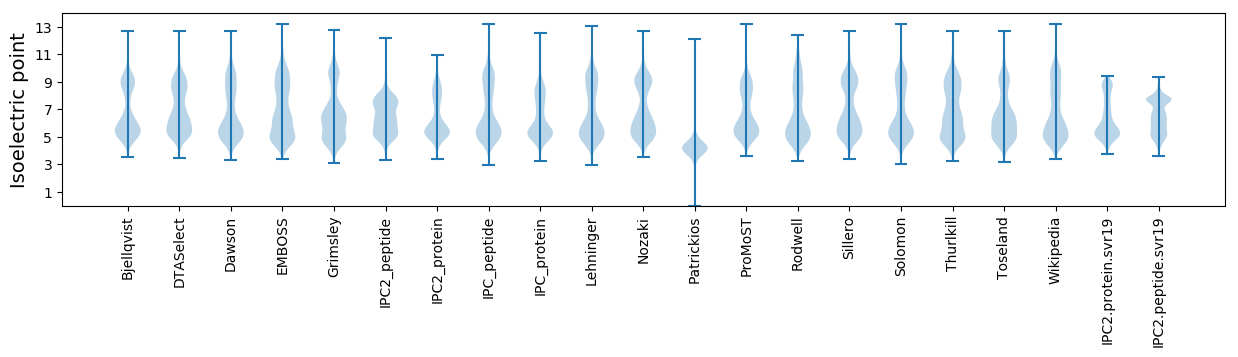
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2X0VTE9|A0A2X0VTE9_9GAMM Transposase OS=Anaerobiospirillum thomasii OX=179995 GN=NCTC13093_02409 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTQKK11 pKa = 10.75RR12 pKa = 11.84KK13 pKa = 7.4RR14 pKa = 11.84THH16 pKa = 5.6GFRR19 pKa = 11.84VRR21 pKa = 11.84MRR23 pKa = 11.84TADD26 pKa = 3.26GRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.88GRR39 pKa = 11.84KK40 pKa = 9.09RR41 pKa = 11.84LTTVNGG47 pKa = 3.51
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNTQKK11 pKa = 10.75RR12 pKa = 11.84KK13 pKa = 7.4RR14 pKa = 11.84THH16 pKa = 5.6GFRR19 pKa = 11.84VRR21 pKa = 11.84MRR23 pKa = 11.84TADD26 pKa = 3.26GRR28 pKa = 11.84KK29 pKa = 8.48VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.88GRR39 pKa = 11.84KK40 pKa = 9.09RR41 pKa = 11.84LTTVNGG47 pKa = 3.51
Molecular weight: 5.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
796788 |
29 |
2574 |
336.5 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.971 ± 0.057 | 1.536 ± 0.02 |
6.334 ± 0.046 | 5.772 ± 0.048 |
4.39 ± 0.034 | 6.143 ± 0.047 |
1.954 ± 0.022 | 7.578 ± 0.042 |
6.791 ± 0.042 | 9.795 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.61 ± 0.024 | 4.97 ± 0.045 |
3.168 ± 0.032 | 3.406 ± 0.032 |
4.386 ± 0.038 | 7.2 ± 0.039 |
5.173 ± 0.032 | 6.497 ± 0.041 |
0.638 ± 0.013 | 3.689 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
