
Candidatus Electrothrix communis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Candidatus Electrothrix
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
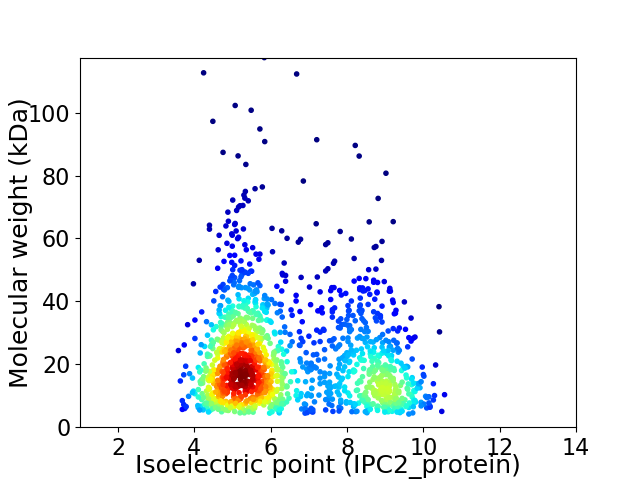
Virtual 2D-PAGE plot for 1454 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S3QYK6|A0A3S3QYK6_9DELT Septal ring factor EnvC activator of murein hydrolases AmiA and AmiB (Fragment) OS=Candidatus Electrothrix communis OX=1859133 GN=VT98_13112 PE=4 SV=1
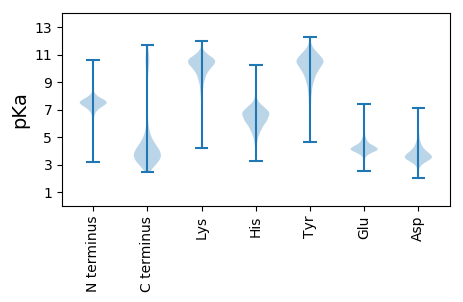
MM1 pKa = 7.19VAGNAQAVEE10 pKa = 3.94HH11 pKa = 6.93RR12 pKa = 11.84IGGGANYY19 pKa = 6.93WTSVDD24 pKa = 6.02DD25 pKa = 4.52IDD27 pKa = 5.88LDD29 pKa = 5.18KK30 pKa = 11.02IDD32 pKa = 5.49DD33 pKa = 4.44DD34 pKa = 4.52GFSFLASYY42 pKa = 8.11QFWPGLIGLEE52 pKa = 4.13LDD54 pKa = 4.02LEE56 pKa = 4.49IVPDD60 pKa = 3.73QYY62 pKa = 11.94GEE64 pKa = 3.91TAYY67 pKa = 10.52APQAYY72 pKa = 9.98ALIGKK77 pKa = 9.43AIYY80 pKa = 9.97AGAGIGINYY89 pKa = 9.31RR90 pKa = 11.84DD91 pKa = 4.01GDD93 pKa = 4.01LADD96 pKa = 4.18EE97 pKa = 4.85PFFSLRR103 pKa = 11.84AGLNLEE109 pKa = 4.3VLPGIYY115 pKa = 9.13WDD117 pKa = 3.09IYY119 pKa = 9.8GNYY122 pKa = 9.22RR123 pKa = 11.84FNDD126 pKa = 3.63TTDD129 pKa = 4.49LEE131 pKa = 4.41DD132 pKa = 3.9HH133 pKa = 6.33STDD136 pKa = 3.39IDD138 pKa = 3.94SDD140 pKa = 4.39TIFLGSAVRR149 pKa = 11.84IAII152 pKa = 4.09
MM1 pKa = 7.19VAGNAQAVEE10 pKa = 3.94HH11 pKa = 6.93RR12 pKa = 11.84IGGGANYY19 pKa = 6.93WTSVDD24 pKa = 6.02DD25 pKa = 4.52IDD27 pKa = 5.88LDD29 pKa = 5.18KK30 pKa = 11.02IDD32 pKa = 5.49DD33 pKa = 4.44DD34 pKa = 4.52GFSFLASYY42 pKa = 8.11QFWPGLIGLEE52 pKa = 4.13LDD54 pKa = 4.02LEE56 pKa = 4.49IVPDD60 pKa = 3.73QYY62 pKa = 11.94GEE64 pKa = 3.91TAYY67 pKa = 10.52APQAYY72 pKa = 9.98ALIGKK77 pKa = 9.43AIYY80 pKa = 9.97AGAGIGINYY89 pKa = 9.31RR90 pKa = 11.84DD91 pKa = 4.01GDD93 pKa = 4.01LADD96 pKa = 4.18EE97 pKa = 4.85PFFSLRR103 pKa = 11.84AGLNLEE109 pKa = 4.3VLPGIYY115 pKa = 9.13WDD117 pKa = 3.09IYY119 pKa = 9.8GNYY122 pKa = 9.22RR123 pKa = 11.84FNDD126 pKa = 3.63TTDD129 pKa = 4.49LEE131 pKa = 4.41DD132 pKa = 3.9HH133 pKa = 6.33STDD136 pKa = 3.39IDD138 pKa = 3.94SDD140 pKa = 4.39TIFLGSAVRR149 pKa = 11.84IAII152 pKa = 4.09
Molecular weight: 16.65 kDa
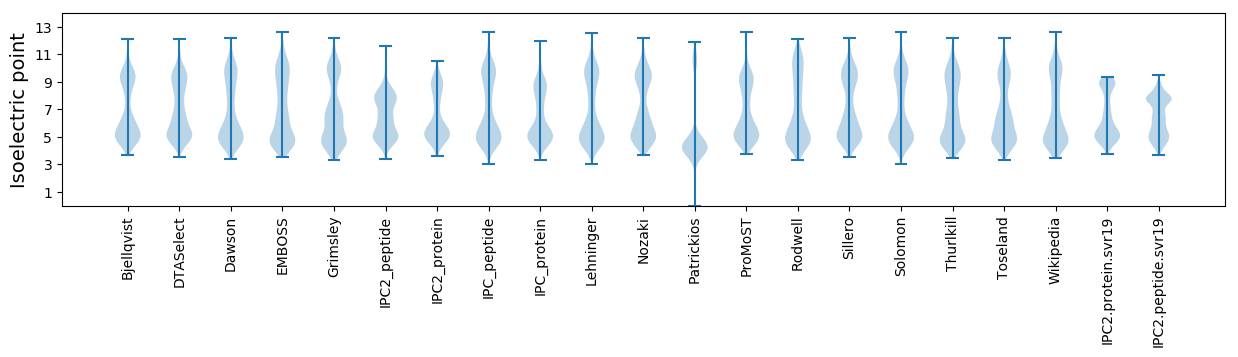
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S3RB49|A0A3S3RB49_9DELT Transcription-repair-coupling factor OS=Candidatus Electrothrix communis OX=1859133 GN=mfd PE=3 SV=1
MM1 pKa = 7.38IRR3 pKa = 11.84GEE5 pKa = 4.34VKK7 pKa = 9.79QQADD11 pKa = 3.31IGTEE15 pKa = 3.5ISGCFQLKK23 pKa = 9.08TGQLQHH29 pKa = 6.55HH30 pKa = 6.71NFRR33 pKa = 11.84FSWIQQIFQNRR44 pKa = 11.84GADD47 pKa = 3.07IAADD51 pKa = 3.62KK52 pKa = 11.19NLLARR57 pKa = 11.84AAQEE61 pKa = 4.01MTDD64 pKa = 3.29QAGYY68 pKa = 10.93GGFAVGPGHH77 pKa = 7.36RR78 pKa = 11.84DD79 pKa = 3.43DD80 pKa = 4.55RR81 pKa = 11.84HH82 pKa = 6.37SDD84 pKa = 3.39PASGEE89 pKa = 4.15GKK91 pKa = 10.35LSNNRR96 pKa = 11.84HH97 pKa = 5.69APSSGRR103 pKa = 11.84SQHH106 pKa = 5.59RR107 pKa = 11.84QIHH110 pKa = 5.48RR111 pKa = 11.84HH112 pKa = 5.26TGTGHH117 pKa = 4.47NQIRR121 pKa = 11.84IDD123 pKa = 3.64KK124 pKa = 9.51KK125 pKa = 8.37GSRR128 pKa = 11.84MAACLQLSAEE138 pKa = 4.69IYY140 pKa = 10.3AALHH144 pKa = 4.67KK145 pKa = 10.47HH146 pKa = 5.33GLLFLRR152 pKa = 11.84DD153 pKa = 4.09RR154 pKa = 11.84LPKK157 pKa = 10.21LRR159 pKa = 11.84NRR161 pKa = 11.84DD162 pKa = 3.37IQSLLVQIFHH172 pKa = 7.03RR173 pKa = 11.84RR174 pKa = 11.84QPGFSQADD182 pKa = 3.49HH183 pKa = 6.86NDD185 pKa = 2.85FHH187 pKa = 8.7ARR189 pKa = 11.84LQFNHH194 pKa = 6.21NLSTEE199 pKa = 4.43IICISGKK206 pKa = 10.44LSAFALLKK214 pKa = 9.42RR215 pKa = 11.84TYY217 pKa = 9.62ILRR220 pKa = 11.84RR221 pKa = 11.84SSVFVTGRR229 pKa = 11.84LFLLYY234 pKa = 10.54CLLLSTNFFIFSRR247 pKa = 11.84RR248 pKa = 11.84ILFYY252 pKa = 11.32NLLLCCNFFHH262 pKa = 7.92LSRR265 pKa = 11.84FVLVYY270 pKa = 10.81SLL272 pKa = 4.19
MM1 pKa = 7.38IRR3 pKa = 11.84GEE5 pKa = 4.34VKK7 pKa = 9.79QQADD11 pKa = 3.31IGTEE15 pKa = 3.5ISGCFQLKK23 pKa = 9.08TGQLQHH29 pKa = 6.55HH30 pKa = 6.71NFRR33 pKa = 11.84FSWIQQIFQNRR44 pKa = 11.84GADD47 pKa = 3.07IAADD51 pKa = 3.62KK52 pKa = 11.19NLLARR57 pKa = 11.84AAQEE61 pKa = 4.01MTDD64 pKa = 3.29QAGYY68 pKa = 10.93GGFAVGPGHH77 pKa = 7.36RR78 pKa = 11.84DD79 pKa = 3.43DD80 pKa = 4.55RR81 pKa = 11.84HH82 pKa = 6.37SDD84 pKa = 3.39PASGEE89 pKa = 4.15GKK91 pKa = 10.35LSNNRR96 pKa = 11.84HH97 pKa = 5.69APSSGRR103 pKa = 11.84SQHH106 pKa = 5.59RR107 pKa = 11.84QIHH110 pKa = 5.48RR111 pKa = 11.84HH112 pKa = 5.26TGTGHH117 pKa = 4.47NQIRR121 pKa = 11.84IDD123 pKa = 3.64KK124 pKa = 9.51KK125 pKa = 8.37GSRR128 pKa = 11.84MAACLQLSAEE138 pKa = 4.69IYY140 pKa = 10.3AALHH144 pKa = 4.67KK145 pKa = 10.47HH146 pKa = 5.33GLLFLRR152 pKa = 11.84DD153 pKa = 4.09RR154 pKa = 11.84LPKK157 pKa = 10.21LRR159 pKa = 11.84NRR161 pKa = 11.84DD162 pKa = 3.37IQSLLVQIFHH172 pKa = 7.03RR173 pKa = 11.84RR174 pKa = 11.84QPGFSQADD182 pKa = 3.49HH183 pKa = 6.86NDD185 pKa = 2.85FHH187 pKa = 8.7ARR189 pKa = 11.84LQFNHH194 pKa = 6.21NLSTEE199 pKa = 4.43IICISGKK206 pKa = 10.44LSAFALLKK214 pKa = 9.42RR215 pKa = 11.84TYY217 pKa = 9.62ILRR220 pKa = 11.84RR221 pKa = 11.84SSVFVTGRR229 pKa = 11.84LFLLYY234 pKa = 10.54CLLLSTNFFIFSRR247 pKa = 11.84RR248 pKa = 11.84ILFYY252 pKa = 11.32NLLLCCNFFHH262 pKa = 7.92LSRR265 pKa = 11.84FVLVYY270 pKa = 10.81SLL272 pKa = 4.19
Molecular weight: 31.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
300695 |
39 |
1036 |
206.8 |
23.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.033 ± 0.075 | 1.384 ± 0.03 |
5.373 ± 0.057 | 6.91 ± 0.071 |
4.24 ± 0.052 | 7.334 ± 0.065 |
2.102 ± 0.033 | 6.276 ± 0.052 |
5.659 ± 0.075 | 10.399 ± 0.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.69 ± 0.035 | 3.616 ± 0.045 |
4.276 ± 0.057 | 4.218 ± 0.045 |
5.546 ± 0.054 | 5.962 ± 0.062 |
5.198 ± 0.056 | 6.856 ± 0.051 |
1.062 ± 0.022 | 2.865 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
