
Salmonella bongori
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales;
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
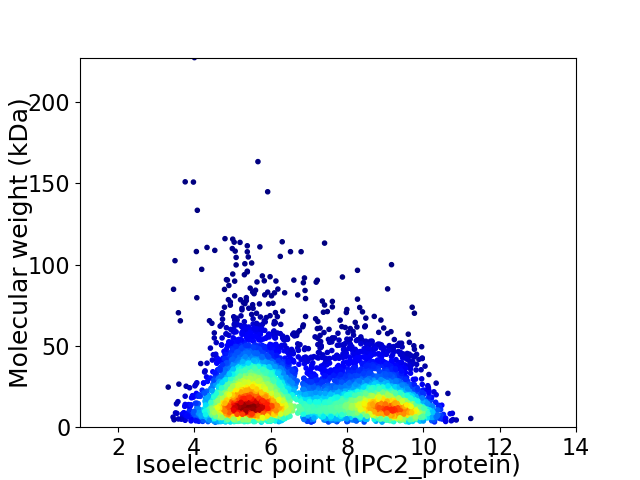
Virtual 2D-PAGE plot for 6192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A447M053|A0A447M053_SALBN Holo-[acyl-carrier-protein] synthase OS=Salmonella bongori OX=54736 GN=acpS PE=3 SV=1
MM1 pKa = 6.79ITVIAIDD8 pKa = 3.47AAGNASPVSDD18 pKa = 3.24SVNFVVDD25 pKa = 3.89TTPPLAPVITSVSDD39 pKa = 3.52DD40 pKa = 3.59QAPGLGTIATGQNTNDD56 pKa = 3.57PTPTFSGTAEE66 pKa = 4.03AGATITLYY74 pKa = 11.12EE75 pKa = 4.43NGTVIGTTTAQPDD88 pKa = 4.42GAWSVPTSTLASGTHH103 pKa = 6.44VITAVATDD111 pKa = 3.5TAGNDD116 pKa = 3.92SPNSTAFTLTVDD128 pKa = 3.85TTAPQTPILTSVVDD142 pKa = 4.27DD143 pKa = 3.81VAGGVTGNLANGQITNDD160 pKa = 3.47NRR162 pKa = 11.84PTLNGTAEE170 pKa = 4.26AGSVVSIYY178 pKa = 11.08DD179 pKa = 3.44GDD181 pKa = 3.95TLLGVTSANTGGAWSFTPTTGLNDD205 pKa = 3.54GTHH208 pKa = 5.22TLKK211 pKa = 9.55VTATDD216 pKa = 3.52PAGNVSPATSGFTIVVDD233 pKa = 3.97TLAPTLPLITSIIDD247 pKa = 3.31DD248 pKa = 3.77VLNNTGAISNGQSTNDD264 pKa = 3.58TQPTLNGTAEE274 pKa = 4.2ANSAVNIFDD283 pKa = 3.8NGTLVATVNASANGNWSWTPTVEE306 pKa = 4.67LGQGSHH312 pKa = 6.9AYY314 pKa = 9.73SVSAADD320 pKa = 3.33AAGNISAASQLTTIIVDD337 pKa = 3.79TLAPGTPGNLVINATGNRR355 pKa = 11.84VTGTAEE361 pKa = 4.09AGSMVTITSDD371 pKa = 3.07TGVVLGTATADD382 pKa = 3.5GTGSFTATLTPAQTNGQPLLAFAQDD407 pKa = 3.34KK408 pKa = 11.01AGNTGIAARR417 pKa = 11.84FTAPDD422 pKa = 3.4TRR424 pKa = 11.84VPEE427 pKa = 4.26APVITNIVDD436 pKa = 3.42DD437 pKa = 4.05VGIYY441 pKa = 9.83TGVIANGQVTNDD453 pKa = 3.81ARR455 pKa = 11.84PTLNGTAQAGATVSIYY471 pKa = 11.32NNGALLGITTANASGNWSFTPTGNLTEE498 pKa = 4.61GSHH501 pKa = 6.73AFTATATNANGTGSVSTAATVMVDD525 pKa = 3.1TLAPGTPSGTLSADD539 pKa = 3.37GGSLSGLAEE548 pKa = 4.49ANSTVTVTLAGGVTLTTTAGSNGAWSLTLPTKK580 pKa = 10.44QIEE583 pKa = 4.44GQLINVTATDD593 pKa = 3.5AAGNASGTLGITAPVLPLAARR614 pKa = 11.84DD615 pKa = 4.2NITSLDD621 pKa = 3.9LTSTAATSTQHH632 pKa = 5.66YY633 pKa = 9.99SDD635 pKa = 4.27YY636 pKa = 11.63GLLLVGALGNVASVLGNDD654 pKa = 3.59TAQVEE659 pKa = 4.44FTIAEE664 pKa = 4.54GGTGDD669 pKa = 3.66VTIDD673 pKa = 3.14AAATGIVLSLLSTQEE688 pKa = 3.94IVVQRR693 pKa = 11.84YY694 pKa = 7.26DD695 pKa = 3.21TSLGAWTTIVNTAVGDD711 pKa = 3.91FANLLTLTGSGVTLNLSGLSEE732 pKa = 3.9GQYY735 pKa = 10.58RR736 pKa = 11.84VLTYY740 pKa = 9.12NTSLLATGSYY750 pKa = 10.1TSLDD754 pKa = 3.32VDD756 pKa = 3.58VHH758 pKa = 4.66QTSAGIISGPTINTGNVMADD778 pKa = 3.28DD779 pKa = 4.18TAPTGTTVTAITNASGISSPVGAGGVDD806 pKa = 2.88IQGQYY811 pKa = 8.41GTLHH815 pKa = 6.56INQDD819 pKa = 2.73GSYY822 pKa = 9.43TYY824 pKa = 10.99TLTNPTAGYY833 pKa = 8.61GHH835 pKa = 7.14KK836 pKa = 10.71EE837 pKa = 3.72NFTYY841 pKa = 10.21TITQNGVGSSTAQLVINLGPAPVPGSVVATDD872 pKa = 3.83NNASLVFDD880 pKa = 3.12THH882 pKa = 7.16VNYY885 pKa = 10.43VNNGPSTQSGVTVLSVGLGNVLSANLLDD913 pKa = 5.77DD914 pKa = 3.61MTNPILFNVEE924 pKa = 3.6EE925 pKa = 4.77GATRR929 pKa = 11.84TLTLQGTVGGVSLVSTFDD947 pKa = 3.41LYY949 pKa = 11.31VYY951 pKa = 10.37RR952 pKa = 11.84FNDD955 pKa = 3.53VIQQYY960 pKa = 8.12EE961 pKa = 3.84QFRR964 pKa = 11.84VQKK967 pKa = 10.52GWINTLLLAGQSQPLTLTLPGGEE990 pKa = 4.13YY991 pKa = 10.81LFVLNTASGISVLTGYY1007 pKa = 8.41TLTVSQDD1014 pKa = 2.51HH1015 pKa = 6.58TYY1017 pKa = 11.3AVDD1020 pKa = 3.51SMTANTTGNVLTNDD1034 pKa = 3.5IAPADD1039 pKa = 3.77ALLTEE1044 pKa = 4.49VNGVAISATGTTEE1057 pKa = 3.84INGLYY1062 pKa = 8.62GTLTIDD1068 pKa = 3.28AKK1070 pKa = 11.3GNYY1073 pKa = 8.46TYY1075 pKa = 10.1TLKK1078 pKa = 11.12NGVGADD1084 pKa = 4.22GIKK1087 pKa = 9.81TPDD1090 pKa = 3.06SFIYY1094 pKa = 9.0TVKK1097 pKa = 10.74APNGDD1102 pKa = 3.43TDD1104 pKa = 3.94TASLNITPTARR1115 pKa = 11.84DD1116 pKa = 3.13LDD1118 pKa = 4.77AINDD1122 pKa = 3.64VSDD1125 pKa = 3.84TLSVATLQDD1134 pKa = 3.04TAAWLDD1140 pKa = 3.64SSVGSASWGLLGKK1153 pKa = 10.03SGSGSGTFDD1162 pKa = 3.08VATGTVLKK1170 pKa = 10.45GASLVFDD1177 pKa = 4.06VSTLITLGNLNISWTIQEE1195 pKa = 4.29NGTVIRR1201 pKa = 11.84NGTVPVANITLGGATVTVNLSGLEE1225 pKa = 4.01LDD1227 pKa = 4.05AGTYY1231 pKa = 7.75TLNFTGTNTLAGAATITPRR1250 pKa = 11.84VIGTTVDD1257 pKa = 3.36LDD1259 pKa = 3.63NFEE1262 pKa = 4.46TSGTHH1267 pKa = 4.84TVPGNIFDD1275 pKa = 4.44GSDD1278 pKa = 2.76AAGAMDD1284 pKa = 4.74QLNTVNTRR1292 pKa = 11.84LSISGYY1298 pKa = 9.16NGSAATLDD1306 pKa = 3.51AAANTTSATIQGHH1319 pKa = 5.86YY1320 pKa = 8.63GTLQINLDD1328 pKa = 3.33GAYY1331 pKa = 9.35TYY1333 pKa = 10.31TLNNGVAISSITHH1346 pKa = 6.16KK1347 pKa = 10.8EE1348 pKa = 3.86VFTYY1352 pKa = 10.71QLDD1355 pKa = 4.04DD1356 pKa = 4.3KK1357 pKa = 11.44NGHH1360 pKa = 5.78TDD1362 pKa = 3.19SATLTIDD1369 pKa = 3.2MAPQIVSTNQNDD1381 pKa = 3.75VLIGSAYY1388 pKa = 10.11GDD1390 pKa = 3.38TLIYY1394 pKa = 10.54HH1395 pKa = 7.46LLNSADD1401 pKa = 3.43ATGGNGADD1409 pKa = 2.93RR1410 pKa = 11.84WQNFSTAQGDD1420 pKa = 3.76KK1421 pKa = 10.55IDD1423 pKa = 3.45IHH1425 pKa = 6.44EE1426 pKa = 5.34LLTGWDD1432 pKa = 3.71HH1433 pKa = 5.81QTATLGNFIQVSTSGANTVISVDD1456 pKa = 3.37RR1457 pKa = 11.84DD1458 pKa = 3.52GAGSAFKK1465 pKa = 10.2ATDD1468 pKa = 3.49LVTLEE1473 pKa = 4.08NVQLSLNDD1481 pKa = 3.86LLQNNHH1487 pKa = 7.48LITGGG1492 pKa = 3.36
MM1 pKa = 6.79ITVIAIDD8 pKa = 3.47AAGNASPVSDD18 pKa = 3.24SVNFVVDD25 pKa = 3.89TTPPLAPVITSVSDD39 pKa = 3.52DD40 pKa = 3.59QAPGLGTIATGQNTNDD56 pKa = 3.57PTPTFSGTAEE66 pKa = 4.03AGATITLYY74 pKa = 11.12EE75 pKa = 4.43NGTVIGTTTAQPDD88 pKa = 4.42GAWSVPTSTLASGTHH103 pKa = 6.44VITAVATDD111 pKa = 3.5TAGNDD116 pKa = 3.92SPNSTAFTLTVDD128 pKa = 3.85TTAPQTPILTSVVDD142 pKa = 4.27DD143 pKa = 3.81VAGGVTGNLANGQITNDD160 pKa = 3.47NRR162 pKa = 11.84PTLNGTAEE170 pKa = 4.26AGSVVSIYY178 pKa = 11.08DD179 pKa = 3.44GDD181 pKa = 3.95TLLGVTSANTGGAWSFTPTTGLNDD205 pKa = 3.54GTHH208 pKa = 5.22TLKK211 pKa = 9.55VTATDD216 pKa = 3.52PAGNVSPATSGFTIVVDD233 pKa = 3.97TLAPTLPLITSIIDD247 pKa = 3.31DD248 pKa = 3.77VLNNTGAISNGQSTNDD264 pKa = 3.58TQPTLNGTAEE274 pKa = 4.2ANSAVNIFDD283 pKa = 3.8NGTLVATVNASANGNWSWTPTVEE306 pKa = 4.67LGQGSHH312 pKa = 6.9AYY314 pKa = 9.73SVSAADD320 pKa = 3.33AAGNISAASQLTTIIVDD337 pKa = 3.79TLAPGTPGNLVINATGNRR355 pKa = 11.84VTGTAEE361 pKa = 4.09AGSMVTITSDD371 pKa = 3.07TGVVLGTATADD382 pKa = 3.5GTGSFTATLTPAQTNGQPLLAFAQDD407 pKa = 3.34KK408 pKa = 11.01AGNTGIAARR417 pKa = 11.84FTAPDD422 pKa = 3.4TRR424 pKa = 11.84VPEE427 pKa = 4.26APVITNIVDD436 pKa = 3.42DD437 pKa = 4.05VGIYY441 pKa = 9.83TGVIANGQVTNDD453 pKa = 3.81ARR455 pKa = 11.84PTLNGTAQAGATVSIYY471 pKa = 11.32NNGALLGITTANASGNWSFTPTGNLTEE498 pKa = 4.61GSHH501 pKa = 6.73AFTATATNANGTGSVSTAATVMVDD525 pKa = 3.1TLAPGTPSGTLSADD539 pKa = 3.37GGSLSGLAEE548 pKa = 4.49ANSTVTVTLAGGVTLTTTAGSNGAWSLTLPTKK580 pKa = 10.44QIEE583 pKa = 4.44GQLINVTATDD593 pKa = 3.5AAGNASGTLGITAPVLPLAARR614 pKa = 11.84DD615 pKa = 4.2NITSLDD621 pKa = 3.9LTSTAATSTQHH632 pKa = 5.66YY633 pKa = 9.99SDD635 pKa = 4.27YY636 pKa = 11.63GLLLVGALGNVASVLGNDD654 pKa = 3.59TAQVEE659 pKa = 4.44FTIAEE664 pKa = 4.54GGTGDD669 pKa = 3.66VTIDD673 pKa = 3.14AAATGIVLSLLSTQEE688 pKa = 3.94IVVQRR693 pKa = 11.84YY694 pKa = 7.26DD695 pKa = 3.21TSLGAWTTIVNTAVGDD711 pKa = 3.91FANLLTLTGSGVTLNLSGLSEE732 pKa = 3.9GQYY735 pKa = 10.58RR736 pKa = 11.84VLTYY740 pKa = 9.12NTSLLATGSYY750 pKa = 10.1TSLDD754 pKa = 3.32VDD756 pKa = 3.58VHH758 pKa = 4.66QTSAGIISGPTINTGNVMADD778 pKa = 3.28DD779 pKa = 4.18TAPTGTTVTAITNASGISSPVGAGGVDD806 pKa = 2.88IQGQYY811 pKa = 8.41GTLHH815 pKa = 6.56INQDD819 pKa = 2.73GSYY822 pKa = 9.43TYY824 pKa = 10.99TLTNPTAGYY833 pKa = 8.61GHH835 pKa = 7.14KK836 pKa = 10.71EE837 pKa = 3.72NFTYY841 pKa = 10.21TITQNGVGSSTAQLVINLGPAPVPGSVVATDD872 pKa = 3.83NNASLVFDD880 pKa = 3.12THH882 pKa = 7.16VNYY885 pKa = 10.43VNNGPSTQSGVTVLSVGLGNVLSANLLDD913 pKa = 5.77DD914 pKa = 3.61MTNPILFNVEE924 pKa = 3.6EE925 pKa = 4.77GATRR929 pKa = 11.84TLTLQGTVGGVSLVSTFDD947 pKa = 3.41LYY949 pKa = 11.31VYY951 pKa = 10.37RR952 pKa = 11.84FNDD955 pKa = 3.53VIQQYY960 pKa = 8.12EE961 pKa = 3.84QFRR964 pKa = 11.84VQKK967 pKa = 10.52GWINTLLLAGQSQPLTLTLPGGEE990 pKa = 4.13YY991 pKa = 10.81LFVLNTASGISVLTGYY1007 pKa = 8.41TLTVSQDD1014 pKa = 2.51HH1015 pKa = 6.58TYY1017 pKa = 11.3AVDD1020 pKa = 3.51SMTANTTGNVLTNDD1034 pKa = 3.5IAPADD1039 pKa = 3.77ALLTEE1044 pKa = 4.49VNGVAISATGTTEE1057 pKa = 3.84INGLYY1062 pKa = 8.62GTLTIDD1068 pKa = 3.28AKK1070 pKa = 11.3GNYY1073 pKa = 8.46TYY1075 pKa = 10.1TLKK1078 pKa = 11.12NGVGADD1084 pKa = 4.22GIKK1087 pKa = 9.81TPDD1090 pKa = 3.06SFIYY1094 pKa = 9.0TVKK1097 pKa = 10.74APNGDD1102 pKa = 3.43TDD1104 pKa = 3.94TASLNITPTARR1115 pKa = 11.84DD1116 pKa = 3.13LDD1118 pKa = 4.77AINDD1122 pKa = 3.64VSDD1125 pKa = 3.84TLSVATLQDD1134 pKa = 3.04TAAWLDD1140 pKa = 3.64SSVGSASWGLLGKK1153 pKa = 10.03SGSGSGTFDD1162 pKa = 3.08VATGTVLKK1170 pKa = 10.45GASLVFDD1177 pKa = 4.06VSTLITLGNLNISWTIQEE1195 pKa = 4.29NGTVIRR1201 pKa = 11.84NGTVPVANITLGGATVTVNLSGLEE1225 pKa = 4.01LDD1227 pKa = 4.05AGTYY1231 pKa = 7.75TLNFTGTNTLAGAATITPRR1250 pKa = 11.84VIGTTVDD1257 pKa = 3.36LDD1259 pKa = 3.63NFEE1262 pKa = 4.46TSGTHH1267 pKa = 4.84TVPGNIFDD1275 pKa = 4.44GSDD1278 pKa = 2.76AAGAMDD1284 pKa = 4.74QLNTVNTRR1292 pKa = 11.84LSISGYY1298 pKa = 9.16NGSAATLDD1306 pKa = 3.51AAANTTSATIQGHH1319 pKa = 5.86YY1320 pKa = 8.63GTLQINLDD1328 pKa = 3.33GAYY1331 pKa = 9.35TYY1333 pKa = 10.31TLNNGVAISSITHH1346 pKa = 6.16KK1347 pKa = 10.8EE1348 pKa = 3.86VFTYY1352 pKa = 10.71QLDD1355 pKa = 4.04DD1356 pKa = 4.3KK1357 pKa = 11.44NGHH1360 pKa = 5.78TDD1362 pKa = 3.19SATLTIDD1369 pKa = 3.2MAPQIVSTNQNDD1381 pKa = 3.75VLIGSAYY1388 pKa = 10.11GDD1390 pKa = 3.38TLIYY1394 pKa = 10.54HH1395 pKa = 7.46LLNSADD1401 pKa = 3.43ATGGNGADD1409 pKa = 2.93RR1410 pKa = 11.84WQNFSTAQGDD1420 pKa = 3.76KK1421 pKa = 10.55IDD1423 pKa = 3.45IHH1425 pKa = 6.44EE1426 pKa = 5.34LLTGWDD1432 pKa = 3.71HH1433 pKa = 5.81QTATLGNFIQVSTSGANTVISVDD1456 pKa = 3.37RR1457 pKa = 11.84DD1458 pKa = 3.52GAGSAFKK1465 pKa = 10.2ATDD1468 pKa = 3.49LVTLEE1473 pKa = 4.08NVQLSLNDD1481 pKa = 3.86LLQNNHH1487 pKa = 7.48LITGGG1492 pKa = 3.36
Molecular weight: 150.96 kDa
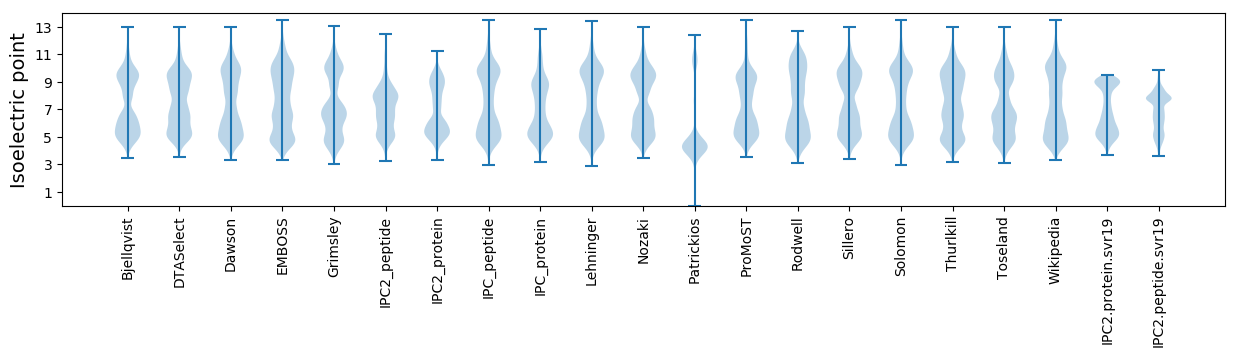
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S5D914|A0A3S5D914_SALBN Glutathione ABC transporter ATP-binding protein OS=Salmonella bongori OX=54736 GN=yheS_3 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208264 |
29 |
2133 |
195.1 |
21.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.629 ± 0.037 | 1.254 ± 0.014 |
5.117 ± 0.029 | 5.395 ± 0.035 |
3.851 ± 0.023 | 7.439 ± 0.031 |
2.334 ± 0.018 | 5.833 ± 0.03 |
4.198 ± 0.03 | 10.545 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.889 ± 0.02 | 3.808 ± 0.025 |
4.569 ± 0.024 | 4.299 ± 0.028 |
5.991 ± 0.037 | 5.894 ± 0.026 |
5.536 ± 0.038 | 6.937 ± 0.03 |
1.579 ± 0.018 | 2.903 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
