
Lachnoclostridium sp. An14
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
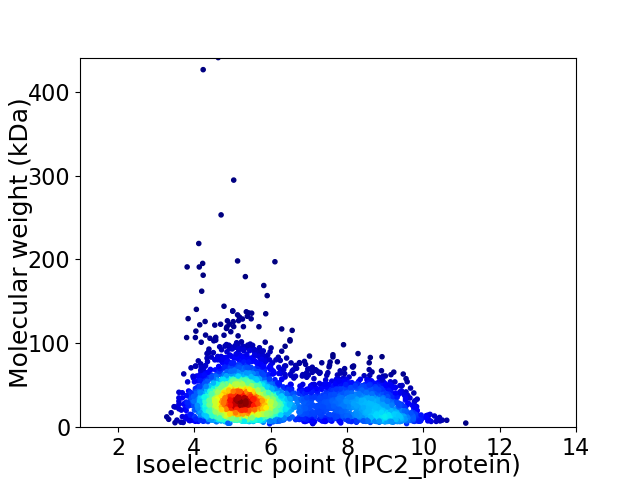
Virtual 2D-PAGE plot for 3897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4RAF8|A0A1Y4RAF8_9FIRM DNA methyltransferase OS=Lachnoclostridium sp. An14 OX=1965562 GN=B5E84_16245 PE=3 SV=1
MM1 pKa = 7.31NMRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.85FFAAVSSLAVTSAVLLTGCGTPAQEE31 pKa = 3.95GTALKK36 pKa = 8.88EE37 pKa = 4.07TGILTLSVNPEE48 pKa = 3.34IQIEE52 pKa = 4.16YY53 pKa = 9.13NQDD56 pKa = 3.14GKK58 pKa = 9.82VTALTGRR65 pKa = 11.84NDD67 pKa = 3.58DD68 pKa = 3.47GKK70 pKa = 11.06GIVEE74 pKa = 4.83MYY76 pKa = 9.99PDD78 pKa = 4.5YY79 pKa = 10.81IGKK82 pKa = 8.87NCDD85 pKa = 3.24DD86 pKa = 3.94VLRR89 pKa = 11.84DD90 pKa = 4.23LIVEE94 pKa = 3.76IHH96 pKa = 5.92EE97 pKa = 4.67AGFFVDD103 pKa = 6.41DD104 pKa = 4.13IDD106 pKa = 4.27GNKK109 pKa = 10.06KK110 pKa = 10.61NIVLQLEE117 pKa = 4.52PGSVLPSNDD126 pKa = 2.94FLADD130 pKa = 3.64MSASTQEE137 pKa = 4.05AVKK140 pKa = 10.22EE141 pKa = 4.04LSLSSGIVTIDD152 pKa = 3.85DD153 pKa = 4.15DD154 pKa = 6.16DD155 pKa = 5.15YY156 pKa = 12.04DD157 pKa = 3.71SAYY160 pKa = 10.72AKK162 pKa = 10.35AGKK165 pKa = 8.81PSPYY169 pKa = 8.73ITLEE173 pKa = 3.83KK174 pKa = 10.31AQEE177 pKa = 3.66IALAQAHH184 pKa = 6.83ISAADD189 pKa = 3.34AVFDD193 pKa = 5.12DD194 pKa = 5.56KK195 pKa = 11.71EE196 pKa = 4.04FDD198 pKa = 4.03HH199 pKa = 7.97DD200 pKa = 4.44DD201 pKa = 3.55GTPIFEE207 pKa = 5.96LEE209 pKa = 4.17FTANGYY215 pKa = 10.28EE216 pKa = 4.03YY217 pKa = 10.11DD218 pKa = 3.96YY219 pKa = 10.75DD220 pKa = 3.4IHH222 pKa = 7.75AVTGKK227 pKa = 9.5VVKK230 pKa = 10.48AEE232 pKa = 4.19HH233 pKa = 6.54KK234 pKa = 10.92AGGAQNEE241 pKa = 4.63SLTTRR246 pKa = 11.84DD247 pKa = 3.45GSTDD251 pKa = 3.64YY252 pKa = 11.61NDD254 pKa = 3.51TDD256 pKa = 3.78YY257 pKa = 11.68GPNNDD262 pKa = 4.65GVTDD266 pKa = 4.02YY267 pKa = 11.63NDD269 pKa = 3.31TDD271 pKa = 3.76YY272 pKa = 11.68GPNNDD277 pKa = 4.65GVTDD281 pKa = 4.02YY282 pKa = 11.63NDD284 pKa = 3.31TDD286 pKa = 3.76YY287 pKa = 11.68GPNNDD292 pKa = 4.65GVTDD296 pKa = 4.02YY297 pKa = 11.79NDD299 pKa = 3.31TDD301 pKa = 4.04YY302 pKa = 12.06GDD304 pKa = 3.65TNYY307 pKa = 11.27DD308 pKa = 4.57DD309 pKa = 6.0GNTDD313 pKa = 3.96YY314 pKa = 11.81GDD316 pKa = 5.6DD317 pKa = 5.49DD318 pKa = 6.7DD319 pKa = 7.7DD320 pKa = 7.02DD321 pKa = 4.56DD322 pKa = 4.53
MM1 pKa = 7.31NMRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.85FFAAVSSLAVTSAVLLTGCGTPAQEE31 pKa = 3.95GTALKK36 pKa = 8.88EE37 pKa = 4.07TGILTLSVNPEE48 pKa = 3.34IQIEE52 pKa = 4.16YY53 pKa = 9.13NQDD56 pKa = 3.14GKK58 pKa = 9.82VTALTGRR65 pKa = 11.84NDD67 pKa = 3.58DD68 pKa = 3.47GKK70 pKa = 11.06GIVEE74 pKa = 4.83MYY76 pKa = 9.99PDD78 pKa = 4.5YY79 pKa = 10.81IGKK82 pKa = 8.87NCDD85 pKa = 3.24DD86 pKa = 3.94VLRR89 pKa = 11.84DD90 pKa = 4.23LIVEE94 pKa = 3.76IHH96 pKa = 5.92EE97 pKa = 4.67AGFFVDD103 pKa = 6.41DD104 pKa = 4.13IDD106 pKa = 4.27GNKK109 pKa = 10.06KK110 pKa = 10.61NIVLQLEE117 pKa = 4.52PGSVLPSNDD126 pKa = 2.94FLADD130 pKa = 3.64MSASTQEE137 pKa = 4.05AVKK140 pKa = 10.22EE141 pKa = 4.04LSLSSGIVTIDD152 pKa = 3.85DD153 pKa = 4.15DD154 pKa = 6.16DD155 pKa = 5.15YY156 pKa = 12.04DD157 pKa = 3.71SAYY160 pKa = 10.72AKK162 pKa = 10.35AGKK165 pKa = 8.81PSPYY169 pKa = 8.73ITLEE173 pKa = 3.83KK174 pKa = 10.31AQEE177 pKa = 3.66IALAQAHH184 pKa = 6.83ISAADD189 pKa = 3.34AVFDD193 pKa = 5.12DD194 pKa = 5.56KK195 pKa = 11.71EE196 pKa = 4.04FDD198 pKa = 4.03HH199 pKa = 7.97DD200 pKa = 4.44DD201 pKa = 3.55GTPIFEE207 pKa = 5.96LEE209 pKa = 4.17FTANGYY215 pKa = 10.28EE216 pKa = 4.03YY217 pKa = 10.11DD218 pKa = 3.96YY219 pKa = 10.75DD220 pKa = 3.4IHH222 pKa = 7.75AVTGKK227 pKa = 9.5VVKK230 pKa = 10.48AEE232 pKa = 4.19HH233 pKa = 6.54KK234 pKa = 10.92AGGAQNEE241 pKa = 4.63SLTTRR246 pKa = 11.84DD247 pKa = 3.45GSTDD251 pKa = 3.64YY252 pKa = 11.61NDD254 pKa = 3.51TDD256 pKa = 3.78YY257 pKa = 11.68GPNNDD262 pKa = 4.65GVTDD266 pKa = 4.02YY267 pKa = 11.63NDD269 pKa = 3.31TDD271 pKa = 3.76YY272 pKa = 11.68GPNNDD277 pKa = 4.65GVTDD281 pKa = 4.02YY282 pKa = 11.63NDD284 pKa = 3.31TDD286 pKa = 3.76YY287 pKa = 11.68GPNNDD292 pKa = 4.65GVTDD296 pKa = 4.02YY297 pKa = 11.79NDD299 pKa = 3.31TDD301 pKa = 4.04YY302 pKa = 12.06GDD304 pKa = 3.65TNYY307 pKa = 11.27DD308 pKa = 4.57DD309 pKa = 6.0GNTDD313 pKa = 3.96YY314 pKa = 11.81GDD316 pKa = 5.6DD317 pKa = 5.49DD318 pKa = 6.7DD319 pKa = 7.7DD320 pKa = 7.02DD321 pKa = 4.56DD322 pKa = 4.53
Molecular weight: 34.94 kDa
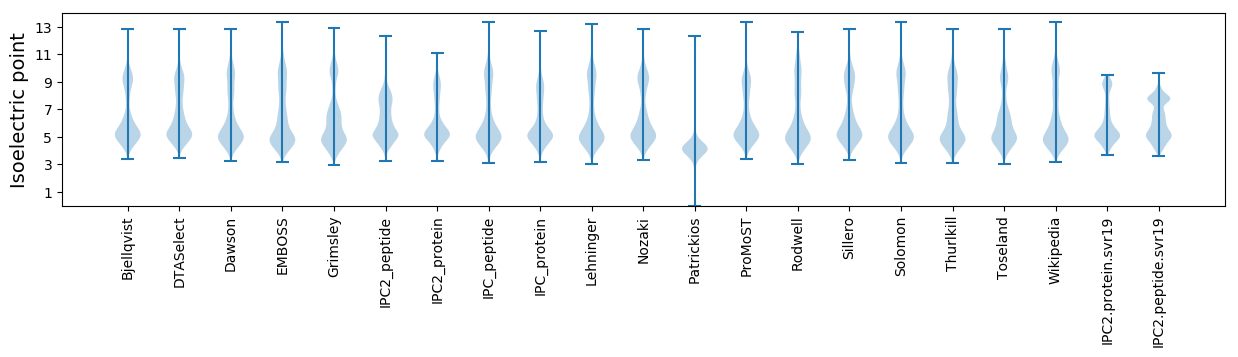
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4R5F5|A0A1Y4R5F5_9FIRM Glutamine-dependent NAD(+) synthetase OS=Lachnoclostridium sp. An14 OX=1965562 GN=nadE PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.57GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.44VHH16 pKa = 5.57GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 4.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1232716 |
34 |
3943 |
316.3 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.565 ± 0.045 | 1.479 ± 0.015 |
5.357 ± 0.035 | 7.829 ± 0.056 |
3.976 ± 0.028 | 8.097 ± 0.045 |
1.668 ± 0.018 | 6.164 ± 0.043 |
5.462 ± 0.037 | 9.324 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.078 ± 0.02 | 3.546 ± 0.023 |
3.847 ± 0.025 | 3.273 ± 0.02 |
5.483 ± 0.042 | 5.57 ± 0.035 |
5.337 ± 0.03 | 7.078 ± 0.031 |
1.056 ± 0.015 | 3.81 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
