
Microviridae sp.
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.99
Get precalculated fractions of proteins
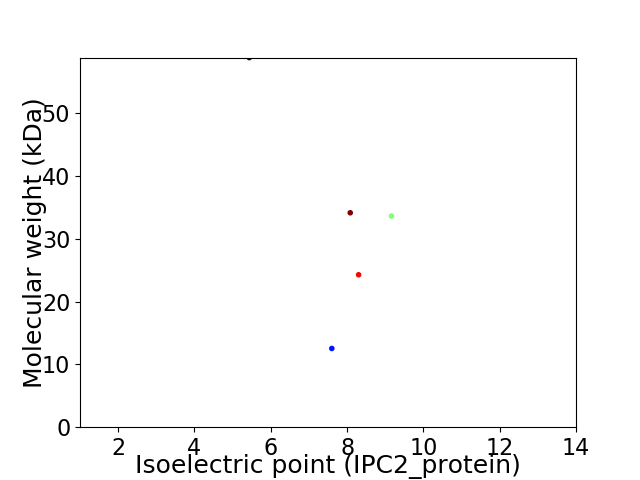
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A345N1A1|A0A345N1A1_9VIRU Major capsid protein OS=Microviridae sp. OX=2202644 PE=3 SV=1
MM1 pKa = 7.63KK2 pKa = 10.52LFDD5 pKa = 3.7QVAVKK10 pKa = 10.32SPKK13 pKa = 9.47YY14 pKa = 10.4SKK16 pKa = 10.72FDD18 pKa = 3.68LSHH21 pKa = 5.89EE22 pKa = 4.5RR23 pKa = 11.84KK24 pKa = 9.79FSMKK28 pKa = 10.17HH29 pKa = 4.1GTLTPIYY36 pKa = 9.11LQEE39 pKa = 4.11VLPGDD44 pKa = 3.97SFRR47 pKa = 11.84LNSEE51 pKa = 3.7MLIRR55 pKa = 11.84LAPMKK60 pKa = 10.55FPVLHH65 pKa = 6.89RR66 pKa = 11.84ISAYY70 pKa = 7.31IHH72 pKa = 5.9YY73 pKa = 9.16WFVPNRR79 pKa = 11.84LVWSEE84 pKa = 3.93WEE86 pKa = 4.12DD87 pKa = 4.53FITGGDD93 pKa = 4.05DD94 pKa = 3.61GLSAPLFPVIDD105 pKa = 4.01EE106 pKa = 4.67TNLTTSQAKK115 pKa = 10.28AGEE118 pKa = 4.27LWDD121 pKa = 4.31FMGMPPIAGTVNPKK135 pKa = 9.57MKK137 pKa = 10.41INALPFRR144 pKa = 11.84GYY146 pKa = 9.21ATIYY150 pKa = 10.34NEE152 pKa = 4.73FYY154 pKa = 10.6RR155 pKa = 11.84DD156 pKa = 3.19QNLQDD161 pKa = 4.8KK162 pKa = 10.91IPVTKK167 pKa = 10.08TGGSLGSAEE176 pKa = 4.07VTEE179 pKa = 4.23LLKK182 pKa = 10.84LRR184 pKa = 11.84YY185 pKa = 9.14RR186 pKa = 11.84AWEE189 pKa = 3.66HH190 pKa = 7.73DD191 pKa = 4.22YY192 pKa = 9.43FTSCLPQAQKK202 pKa = 10.86GGEE205 pKa = 4.04VLLPMEE211 pKa = 5.03ADD213 pKa = 2.92INYY216 pKa = 9.8KK217 pKa = 10.11SEE219 pKa = 3.54ASIRR223 pKa = 11.84MATSSAGYY231 pKa = 8.35TEE233 pKa = 4.4GANVQIQQTGFTPPTAGLFGMEE255 pKa = 4.26GSTARR260 pKa = 11.84AFGIEE265 pKa = 4.21NIEE268 pKa = 4.56SIDD271 pKa = 4.03NATTTINDD279 pKa = 2.88LRR281 pKa = 11.84KK282 pKa = 9.59AVRR285 pKa = 11.84LQEE288 pKa = 4.12FLEE291 pKa = 4.19KK292 pKa = 10.71AMRR295 pKa = 11.84AGSRR299 pKa = 11.84YY300 pKa = 9.49SEE302 pKa = 4.35SIKK305 pKa = 10.64AYY307 pKa = 9.99FGVRR311 pKa = 11.84SSDD314 pKa = 3.12ARR316 pKa = 11.84LQRR319 pKa = 11.84PEE321 pKa = 3.79FLGGGVQPIVVSEE334 pKa = 3.99VLQTYY339 pKa = 9.8EE340 pKa = 4.06GTDD343 pKa = 3.54PLGKK347 pKa = 9.56MGGHH351 pKa = 7.38GISVGNKK358 pKa = 8.83SGFKK362 pKa = 10.26KK363 pKa = 10.25RR364 pKa = 11.84FEE366 pKa = 3.83EE367 pKa = 3.73HH368 pKa = 6.69GYY370 pKa = 10.3IIGIMSIIPKK380 pKa = 9.51SAYY383 pKa = 9.56GQGIPKK389 pKa = 9.19TFMKK393 pKa = 10.55ANRR396 pKa = 11.84YY397 pKa = 9.55DD398 pKa = 3.43FAFPEE403 pKa = 4.77FAQLGEE409 pKa = 4.09QEE411 pKa = 4.43VLNKK415 pKa = 10.06EE416 pKa = 4.5VFWDD420 pKa = 3.68EE421 pKa = 3.92TDD423 pKa = 4.29VNNVWNDD430 pKa = 3.28LEE432 pKa = 4.23FGYY435 pKa = 10.27IPRR438 pKa = 11.84YY439 pKa = 9.74SEE441 pKa = 4.12YY442 pKa = 10.8KK443 pKa = 9.56FNNSSVHH450 pKa = 4.71GQFRR454 pKa = 11.84PGEE457 pKa = 4.13TLEE460 pKa = 3.76NWTMVRR466 pKa = 11.84KK467 pKa = 10.02FEE469 pKa = 4.53NRR471 pKa = 11.84PALNEE476 pKa = 3.93EE477 pKa = 4.79FISSEE482 pKa = 4.77DD483 pKa = 3.25ATQDD487 pKa = 2.92IFNVTDD493 pKa = 4.06PNEE496 pKa = 4.13DD497 pKa = 3.45KK498 pKa = 11.38YY499 pKa = 11.18FVQIYY504 pKa = 10.02HH505 pKa = 6.75NISAIRR511 pKa = 11.84ALPYY515 pKa = 10.48FGTPTLL521 pKa = 3.89
MM1 pKa = 7.63KK2 pKa = 10.52LFDD5 pKa = 3.7QVAVKK10 pKa = 10.32SPKK13 pKa = 9.47YY14 pKa = 10.4SKK16 pKa = 10.72FDD18 pKa = 3.68LSHH21 pKa = 5.89EE22 pKa = 4.5RR23 pKa = 11.84KK24 pKa = 9.79FSMKK28 pKa = 10.17HH29 pKa = 4.1GTLTPIYY36 pKa = 9.11LQEE39 pKa = 4.11VLPGDD44 pKa = 3.97SFRR47 pKa = 11.84LNSEE51 pKa = 3.7MLIRR55 pKa = 11.84LAPMKK60 pKa = 10.55FPVLHH65 pKa = 6.89RR66 pKa = 11.84ISAYY70 pKa = 7.31IHH72 pKa = 5.9YY73 pKa = 9.16WFVPNRR79 pKa = 11.84LVWSEE84 pKa = 3.93WEE86 pKa = 4.12DD87 pKa = 4.53FITGGDD93 pKa = 4.05DD94 pKa = 3.61GLSAPLFPVIDD105 pKa = 4.01EE106 pKa = 4.67TNLTTSQAKK115 pKa = 10.28AGEE118 pKa = 4.27LWDD121 pKa = 4.31FMGMPPIAGTVNPKK135 pKa = 9.57MKK137 pKa = 10.41INALPFRR144 pKa = 11.84GYY146 pKa = 9.21ATIYY150 pKa = 10.34NEE152 pKa = 4.73FYY154 pKa = 10.6RR155 pKa = 11.84DD156 pKa = 3.19QNLQDD161 pKa = 4.8KK162 pKa = 10.91IPVTKK167 pKa = 10.08TGGSLGSAEE176 pKa = 4.07VTEE179 pKa = 4.23LLKK182 pKa = 10.84LRR184 pKa = 11.84YY185 pKa = 9.14RR186 pKa = 11.84AWEE189 pKa = 3.66HH190 pKa = 7.73DD191 pKa = 4.22YY192 pKa = 9.43FTSCLPQAQKK202 pKa = 10.86GGEE205 pKa = 4.04VLLPMEE211 pKa = 5.03ADD213 pKa = 2.92INYY216 pKa = 9.8KK217 pKa = 10.11SEE219 pKa = 3.54ASIRR223 pKa = 11.84MATSSAGYY231 pKa = 8.35TEE233 pKa = 4.4GANVQIQQTGFTPPTAGLFGMEE255 pKa = 4.26GSTARR260 pKa = 11.84AFGIEE265 pKa = 4.21NIEE268 pKa = 4.56SIDD271 pKa = 4.03NATTTINDD279 pKa = 2.88LRR281 pKa = 11.84KK282 pKa = 9.59AVRR285 pKa = 11.84LQEE288 pKa = 4.12FLEE291 pKa = 4.19KK292 pKa = 10.71AMRR295 pKa = 11.84AGSRR299 pKa = 11.84YY300 pKa = 9.49SEE302 pKa = 4.35SIKK305 pKa = 10.64AYY307 pKa = 9.99FGVRR311 pKa = 11.84SSDD314 pKa = 3.12ARR316 pKa = 11.84LQRR319 pKa = 11.84PEE321 pKa = 3.79FLGGGVQPIVVSEE334 pKa = 3.99VLQTYY339 pKa = 9.8EE340 pKa = 4.06GTDD343 pKa = 3.54PLGKK347 pKa = 9.56MGGHH351 pKa = 7.38GISVGNKK358 pKa = 8.83SGFKK362 pKa = 10.26KK363 pKa = 10.25RR364 pKa = 11.84FEE366 pKa = 3.83EE367 pKa = 3.73HH368 pKa = 6.69GYY370 pKa = 10.3IIGIMSIIPKK380 pKa = 9.51SAYY383 pKa = 9.56GQGIPKK389 pKa = 9.19TFMKK393 pKa = 10.55ANRR396 pKa = 11.84YY397 pKa = 9.55DD398 pKa = 3.43FAFPEE403 pKa = 4.77FAQLGEE409 pKa = 4.09QEE411 pKa = 4.43VLNKK415 pKa = 10.06EE416 pKa = 4.5VFWDD420 pKa = 3.68EE421 pKa = 3.92TDD423 pKa = 4.29VNNVWNDD430 pKa = 3.28LEE432 pKa = 4.23FGYY435 pKa = 10.27IPRR438 pKa = 11.84YY439 pKa = 9.74SEE441 pKa = 4.12YY442 pKa = 10.8KK443 pKa = 9.56FNNSSVHH450 pKa = 4.71GQFRR454 pKa = 11.84PGEE457 pKa = 4.13TLEE460 pKa = 3.76NWTMVRR466 pKa = 11.84KK467 pKa = 10.02FEE469 pKa = 4.53NRR471 pKa = 11.84PALNEE476 pKa = 3.93EE477 pKa = 4.79FISSEE482 pKa = 4.77DD483 pKa = 3.25ATQDD487 pKa = 2.92IFNVTDD493 pKa = 4.06PNEE496 pKa = 4.13DD497 pKa = 3.45KK498 pKa = 11.38YY499 pKa = 11.18FVQIYY504 pKa = 10.02HH505 pKa = 6.75NISAIRR511 pKa = 11.84ALPYY515 pKa = 10.48FGTPTLL521 pKa = 3.89
Molecular weight: 58.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345N1A1|A0A345N1A1_9VIRU Major capsid protein OS=Microviridae sp. OX=2202644 PE=3 SV=1
MM1 pKa = 7.29ACEE4 pKa = 4.01RR5 pKa = 11.84PIILKK10 pKa = 10.33DD11 pKa = 3.3HH12 pKa = 6.69LTTVPCRR19 pKa = 11.84RR20 pKa = 11.84CIFCLQEE27 pKa = 5.46KK28 pKa = 9.61RR29 pKa = 11.84DD30 pKa = 3.4NWTFRR35 pKa = 11.84VGQEE39 pKa = 3.77YY40 pKa = 10.64KK41 pKa = 10.42KK42 pKa = 10.98ARR44 pKa = 11.84TAYY47 pKa = 10.26FITLTYY53 pKa = 10.67DD54 pKa = 3.66DD55 pKa = 3.76QHH57 pKa = 8.61LPIDD61 pKa = 4.15NKK63 pKa = 10.35GRR65 pKa = 11.84AYY67 pKa = 10.78LDD69 pKa = 3.23KK70 pKa = 10.55KK71 pKa = 10.53QLVGFLKK78 pKa = 10.49RR79 pKa = 11.84LRR81 pKa = 11.84IRR83 pKa = 11.84NEE85 pKa = 3.79RR86 pKa = 11.84EE87 pKa = 3.3IEE89 pKa = 4.17KK90 pKa = 10.27YY91 pKa = 8.47WRR93 pKa = 11.84ARR95 pKa = 11.84EE96 pKa = 3.91NSTPMPKK103 pKa = 9.0MAKK106 pKa = 8.59TKK108 pKa = 10.46YY109 pKa = 8.97FACGEE114 pKa = 4.14YY115 pKa = 10.53GSKK118 pKa = 10.2KK119 pKa = 9.72KK120 pKa = 10.24RR121 pKa = 11.84PHH123 pKa = 4.34YY124 pKa = 10.22HH125 pKa = 6.31IIVFNLFPNTLKK137 pKa = 10.83YY138 pKa = 10.88LNEE141 pKa = 3.97LWTEE145 pKa = 4.05GNIDD149 pKa = 4.22LKK151 pKa = 10.58IAKK154 pKa = 9.59AGAIHH159 pKa = 5.6YY160 pKa = 7.72TSGYY164 pKa = 9.87LLKK167 pKa = 9.37QTAVIKK173 pKa = 10.46DD174 pKa = 2.82GHH176 pKa = 6.06ARR178 pKa = 11.84QFIIASQHH186 pKa = 5.13MGEE189 pKa = 4.89HH190 pKa = 5.44YY191 pKa = 10.51LKK193 pKa = 10.46NAMYY197 pKa = 10.34HH198 pKa = 5.69SEE200 pKa = 4.21NLTAKK205 pKa = 10.41CSINGFSIGLPSNFRR220 pKa = 11.84NKK222 pKa = 9.82IFTKK226 pKa = 7.62EE227 pKa = 3.65QKK229 pKa = 10.49EE230 pKa = 3.92IMRR233 pKa = 11.84NEE235 pKa = 3.95NEE237 pKa = 4.32KK238 pKa = 10.75KK239 pKa = 10.29RR240 pKa = 11.84LEE242 pKa = 4.57KK243 pKa = 10.12DD244 pKa = 3.73TIWIDD249 pKa = 3.29QKK251 pKa = 9.88TKK253 pKa = 11.04EE254 pKa = 4.57NIDD257 pKa = 3.27FDD259 pKa = 4.72KK260 pKa = 11.5YY261 pKa = 8.05QTEE264 pKa = 4.2QKK266 pKa = 11.0NLLRR270 pKa = 11.84EE271 pKa = 4.13NANKK275 pKa = 8.63KK276 pKa = 7.24TSKK279 pKa = 9.94KK280 pKa = 10.2RR281 pKa = 11.84KK282 pKa = 8.82LL283 pKa = 3.42
MM1 pKa = 7.29ACEE4 pKa = 4.01RR5 pKa = 11.84PIILKK10 pKa = 10.33DD11 pKa = 3.3HH12 pKa = 6.69LTTVPCRR19 pKa = 11.84RR20 pKa = 11.84CIFCLQEE27 pKa = 5.46KK28 pKa = 9.61RR29 pKa = 11.84DD30 pKa = 3.4NWTFRR35 pKa = 11.84VGQEE39 pKa = 3.77YY40 pKa = 10.64KK41 pKa = 10.42KK42 pKa = 10.98ARR44 pKa = 11.84TAYY47 pKa = 10.26FITLTYY53 pKa = 10.67DD54 pKa = 3.66DD55 pKa = 3.76QHH57 pKa = 8.61LPIDD61 pKa = 4.15NKK63 pKa = 10.35GRR65 pKa = 11.84AYY67 pKa = 10.78LDD69 pKa = 3.23KK70 pKa = 10.55KK71 pKa = 10.53QLVGFLKK78 pKa = 10.49RR79 pKa = 11.84LRR81 pKa = 11.84IRR83 pKa = 11.84NEE85 pKa = 3.79RR86 pKa = 11.84EE87 pKa = 3.3IEE89 pKa = 4.17KK90 pKa = 10.27YY91 pKa = 8.47WRR93 pKa = 11.84ARR95 pKa = 11.84EE96 pKa = 3.91NSTPMPKK103 pKa = 9.0MAKK106 pKa = 8.59TKK108 pKa = 10.46YY109 pKa = 8.97FACGEE114 pKa = 4.14YY115 pKa = 10.53GSKK118 pKa = 10.2KK119 pKa = 9.72KK120 pKa = 10.24RR121 pKa = 11.84PHH123 pKa = 4.34YY124 pKa = 10.22HH125 pKa = 6.31IIVFNLFPNTLKK137 pKa = 10.83YY138 pKa = 10.88LNEE141 pKa = 3.97LWTEE145 pKa = 4.05GNIDD149 pKa = 4.22LKK151 pKa = 10.58IAKK154 pKa = 9.59AGAIHH159 pKa = 5.6YY160 pKa = 7.72TSGYY164 pKa = 9.87LLKK167 pKa = 9.37QTAVIKK173 pKa = 10.46DD174 pKa = 2.82GHH176 pKa = 6.06ARR178 pKa = 11.84QFIIASQHH186 pKa = 5.13MGEE189 pKa = 4.89HH190 pKa = 5.44YY191 pKa = 10.51LKK193 pKa = 10.46NAMYY197 pKa = 10.34HH198 pKa = 5.69SEE200 pKa = 4.21NLTAKK205 pKa = 10.41CSINGFSIGLPSNFRR220 pKa = 11.84NKK222 pKa = 9.82IFTKK226 pKa = 7.62EE227 pKa = 3.65QKK229 pKa = 10.49EE230 pKa = 3.92IMRR233 pKa = 11.84NEE235 pKa = 3.95NEE237 pKa = 4.32KK238 pKa = 10.75KK239 pKa = 10.29RR240 pKa = 11.84LEE242 pKa = 4.57KK243 pKa = 10.12DD244 pKa = 3.73TIWIDD249 pKa = 3.29QKK251 pKa = 9.88TKK253 pKa = 11.04EE254 pKa = 4.57NIDD257 pKa = 3.27FDD259 pKa = 4.72KK260 pKa = 11.5YY261 pKa = 8.05QTEE264 pKa = 4.2QKK266 pKa = 11.0NLLRR270 pKa = 11.84EE271 pKa = 4.13NANKK275 pKa = 8.63KK276 pKa = 7.24TSKK279 pKa = 9.94KK280 pKa = 10.2RR281 pKa = 11.84KK282 pKa = 8.82LL283 pKa = 3.42
Molecular weight: 33.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1422 |
107 |
521 |
284.4 |
32.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.048 ± 0.924 | 1.055 ± 0.608 |
4.712 ± 0.472 | 7.314 ± 0.659 |
4.782 ± 0.783 | 6.399 ± 1.063 |
2.25 ± 0.576 | 7.736 ± 0.768 |
8.509 ± 1.497 | 9.705 ± 2.142 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.883 ± 0.408 | 6.188 ± 0.811 |
3.797 ± 0.692 | 4.993 ± 0.692 |
5.345 ± 0.552 | 4.712 ± 0.82 |
4.993 ± 0.919 | 3.797 ± 0.718 |
1.336 ± 0.202 | 3.446 ± 0.667 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
