
Macaca nemestrina rhadinovirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus; Macacine gammaherpesvirus 12
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
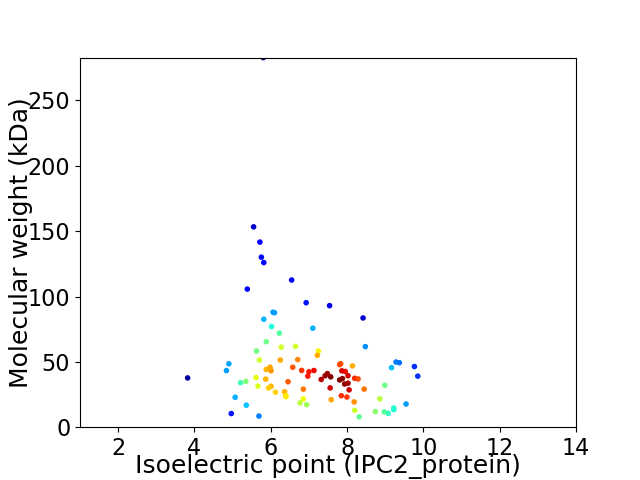
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

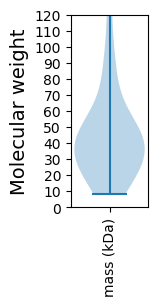
Protein with the lowest isoelectric point:
>tr|A0A0B5CYF8|A0A0B5CYF8_9GAMA ORF49 OS=Macaca nemestrina rhadinovirus 2 OX=123630 GN=ORF49 PE=4 SV=1
MM1 pKa = 7.9AMFLSDD7 pKa = 4.33PPRR10 pKa = 11.84TPPATPRR17 pKa = 11.84MLPIPGAPKK26 pKa = 9.82KK27 pKa = 9.09RR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84FLFAGAGTALPVPPGYY47 pKa = 10.1QGPRR51 pKa = 11.84VIDD54 pKa = 3.3MSAPDD59 pKa = 3.91SVFDD63 pKa = 4.91LDD65 pKa = 5.44SPPTTPKK72 pKa = 10.49TPDD75 pKa = 3.2EE76 pKa = 4.19TDD78 pKa = 3.13SRR80 pKa = 11.84SEE82 pKa = 3.87DD83 pKa = 3.52SNYY86 pKa = 10.75DD87 pKa = 3.43DD88 pKa = 5.41SDD90 pKa = 3.75VDD92 pKa = 3.74EE93 pKa = 4.79EE94 pKa = 5.09TNTEE98 pKa = 4.24PPVSSPPKK106 pKa = 9.67IGLALSEE113 pKa = 4.37GDD115 pKa = 4.21LFRR118 pKa = 11.84PSNTRR123 pKa = 11.84PSVIVEE129 pKa = 4.35SGPVAQPNAPAPLTAFGGPRR149 pKa = 11.84PVAVVTGQHH158 pKa = 6.35RR159 pKa = 11.84APPSSDD165 pKa = 3.45SNSDD169 pKa = 3.36DD170 pKa = 4.48FFIDD174 pKa = 4.09DD175 pKa = 4.5YY176 pKa = 11.62EE177 pKa = 6.35DD178 pKa = 3.92NDD180 pKa = 3.91EE181 pKa = 4.95SDD183 pKa = 4.91NDD185 pKa = 4.14DD186 pKa = 3.71EE187 pKa = 4.92TDD189 pKa = 3.36GFSPRR194 pKa = 11.84GSTGPWPGDD203 pKa = 3.44VSRR206 pKa = 11.84SPPEE210 pKa = 4.42GDD212 pKa = 2.92WSSDD216 pKa = 3.41DD217 pKa = 4.93DD218 pKa = 3.96LTEE221 pKa = 3.98EE222 pKa = 4.03TGAAAGEE229 pKa = 4.03EE230 pKa = 4.31TIIISSSDD238 pKa = 3.96DD239 pKa = 3.61EE240 pKa = 6.71DD241 pKa = 5.37EE242 pKa = 5.68DD243 pKa = 4.17DD244 pKa = 3.92QNSVGTEE251 pKa = 3.91DD252 pKa = 4.96EE253 pKa = 4.13FDD255 pKa = 3.89ANAGSGGADD264 pKa = 3.46VIDD267 pKa = 4.57LCSSSDD273 pKa = 3.5SDD275 pKa = 5.13DD276 pKa = 3.96EE277 pKa = 4.41VDD279 pKa = 3.75GAVGGARR286 pKa = 11.84ANCKK290 pKa = 9.71RR291 pKa = 11.84RR292 pKa = 11.84ASRR295 pKa = 11.84LDD297 pKa = 3.98DD298 pKa = 4.85DD299 pKa = 5.05SEE301 pKa = 5.72DD302 pKa = 4.32DD303 pKa = 3.75IIYY306 pKa = 10.83VGTTQGRR313 pKa = 11.84KK314 pKa = 9.15RR315 pKa = 11.84RR316 pKa = 11.84VTSTAKK322 pKa = 10.63GGATSSSEE330 pKa = 3.94GAGVSGRR337 pKa = 11.84QNMAATPPVCGNDD350 pKa = 3.26NYY352 pKa = 10.38PWPWLDD358 pKa = 3.15
MM1 pKa = 7.9AMFLSDD7 pKa = 4.33PPRR10 pKa = 11.84TPPATPRR17 pKa = 11.84MLPIPGAPKK26 pKa = 9.82KK27 pKa = 9.09RR28 pKa = 11.84TRR30 pKa = 11.84RR31 pKa = 11.84FLFAGAGTALPVPPGYY47 pKa = 10.1QGPRR51 pKa = 11.84VIDD54 pKa = 3.3MSAPDD59 pKa = 3.91SVFDD63 pKa = 4.91LDD65 pKa = 5.44SPPTTPKK72 pKa = 10.49TPDD75 pKa = 3.2EE76 pKa = 4.19TDD78 pKa = 3.13SRR80 pKa = 11.84SEE82 pKa = 3.87DD83 pKa = 3.52SNYY86 pKa = 10.75DD87 pKa = 3.43DD88 pKa = 5.41SDD90 pKa = 3.75VDD92 pKa = 3.74EE93 pKa = 4.79EE94 pKa = 5.09TNTEE98 pKa = 4.24PPVSSPPKK106 pKa = 9.67IGLALSEE113 pKa = 4.37GDD115 pKa = 4.21LFRR118 pKa = 11.84PSNTRR123 pKa = 11.84PSVIVEE129 pKa = 4.35SGPVAQPNAPAPLTAFGGPRR149 pKa = 11.84PVAVVTGQHH158 pKa = 6.35RR159 pKa = 11.84APPSSDD165 pKa = 3.45SNSDD169 pKa = 3.36DD170 pKa = 4.48FFIDD174 pKa = 4.09DD175 pKa = 4.5YY176 pKa = 11.62EE177 pKa = 6.35DD178 pKa = 3.92NDD180 pKa = 3.91EE181 pKa = 4.95SDD183 pKa = 4.91NDD185 pKa = 4.14DD186 pKa = 3.71EE187 pKa = 4.92TDD189 pKa = 3.36GFSPRR194 pKa = 11.84GSTGPWPGDD203 pKa = 3.44VSRR206 pKa = 11.84SPPEE210 pKa = 4.42GDD212 pKa = 2.92WSSDD216 pKa = 3.41DD217 pKa = 4.93DD218 pKa = 3.96LTEE221 pKa = 3.98EE222 pKa = 4.03TGAAAGEE229 pKa = 4.03EE230 pKa = 4.31TIIISSSDD238 pKa = 3.96DD239 pKa = 3.61EE240 pKa = 6.71DD241 pKa = 5.37EE242 pKa = 5.68DD243 pKa = 4.17DD244 pKa = 3.92QNSVGTEE251 pKa = 3.91DD252 pKa = 4.96EE253 pKa = 4.13FDD255 pKa = 3.89ANAGSGGADD264 pKa = 3.46VIDD267 pKa = 4.57LCSSSDD273 pKa = 3.5SDD275 pKa = 5.13DD276 pKa = 3.96EE277 pKa = 4.41VDD279 pKa = 3.75GAVGGARR286 pKa = 11.84ANCKK290 pKa = 9.71RR291 pKa = 11.84RR292 pKa = 11.84ASRR295 pKa = 11.84LDD297 pKa = 3.98DD298 pKa = 4.85DD299 pKa = 5.05SEE301 pKa = 5.72DD302 pKa = 4.32DD303 pKa = 3.75IIYY306 pKa = 10.83VGTTQGRR313 pKa = 11.84KK314 pKa = 9.15RR315 pKa = 11.84RR316 pKa = 11.84VTSTAKK322 pKa = 10.63GGATSSSEE330 pKa = 3.94GAGVSGRR337 pKa = 11.84QNMAATPPVCGNDD350 pKa = 3.26NYY352 pKa = 10.38PWPWLDD358 pKa = 3.15
Molecular weight: 37.69 kDa
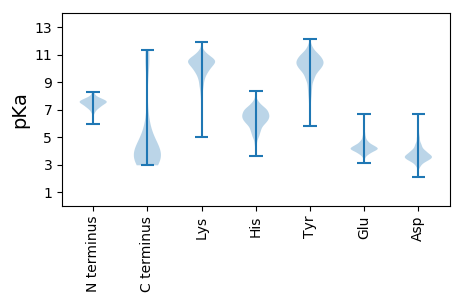
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5D3J3|A0A0B5D3J3_9GAMA Major capsid protein OS=Macaca nemestrina rhadinovirus 2 OX=123630 GN=ORF25 PE=3 SV=1
MM1 pKa = 7.48SLDD4 pKa = 3.4NQHH7 pKa = 6.07RR8 pKa = 11.84HH9 pKa = 4.71SAALEE14 pKa = 4.0CLPTARR20 pKa = 11.84KK21 pKa = 9.03RR22 pKa = 11.84AGIRR26 pKa = 11.84AHH28 pKa = 5.85LAVYY32 pKa = 10.09RR33 pKa = 11.84RR34 pKa = 11.84LIKK37 pKa = 10.3HH38 pKa = 6.52RR39 pKa = 11.84SLGDD43 pKa = 2.99IFKK46 pKa = 10.49FLSICSTRR54 pKa = 11.84EE55 pKa = 3.79ATEE58 pKa = 4.05DD59 pKa = 3.15AQFRR63 pKa = 11.84IFFEE67 pKa = 4.12VTLGKK72 pKa = 10.47RR73 pKa = 11.84IADD76 pKa = 3.44CVLTVEE82 pKa = 4.97SKK84 pKa = 10.16NQKK87 pKa = 6.26TCYY90 pKa = 9.65VVEE93 pKa = 4.86LKK95 pKa = 9.6TCLSGFVFPGSAIKK109 pKa = 10.43ISQRR113 pKa = 11.84RR114 pKa = 11.84QGLEE118 pKa = 3.57QLTDD122 pKa = 3.31SVAYY126 pKa = 9.51IGRR129 pKa = 11.84AAPKK133 pKa = 10.09GHH135 pKa = 6.51EE136 pKa = 4.15NWSVRR141 pKa = 11.84PLLLFKK147 pKa = 10.55NQKK150 pKa = 5.53TLKK153 pKa = 8.91TIHH156 pKa = 6.4TEE158 pKa = 3.89SSAFPPTFINTTSVALNGFFNQWEE182 pKa = 4.24DD183 pKa = 3.25ARR185 pKa = 11.84VRR187 pKa = 11.84KK188 pKa = 8.62MLYY191 pKa = 9.97AIPAKK196 pKa = 9.58THH198 pKa = 5.32ATNNLEE204 pKa = 4.21FLDD207 pKa = 4.34PASKK211 pKa = 10.09QRR213 pKa = 11.84SVYY216 pKa = 10.4SQTIVNGGKK225 pKa = 9.91KK226 pKa = 9.79KK227 pKa = 10.08RR228 pKa = 11.84VRR230 pKa = 11.84DD231 pKa = 3.75AKK233 pKa = 10.05STAGAQGSRR242 pKa = 11.84AEE244 pKa = 4.74KK245 pKa = 10.38KK246 pKa = 9.19PAPARR251 pKa = 11.84ARR253 pKa = 11.84QRR255 pKa = 11.84AANAPTGNGDD265 pKa = 3.35GYY267 pKa = 11.32ARR269 pKa = 11.84HH270 pKa = 6.62RR271 pKa = 11.84DD272 pKa = 3.69DD273 pKa = 4.01PRR275 pKa = 11.84HH276 pKa = 5.03GRR278 pKa = 11.84RR279 pKa = 11.84FASTEE284 pKa = 3.51QKK286 pKa = 9.74PRR288 pKa = 11.84RR289 pKa = 11.84NRR291 pKa = 11.84TVNRR295 pKa = 11.84PATQNRR301 pKa = 11.84PSDD304 pKa = 2.92AWRR307 pKa = 11.84HH308 pKa = 4.11VRR310 pKa = 11.84GHH312 pKa = 5.96NSPRR316 pKa = 11.84GRR318 pKa = 11.84GFYY321 pKa = 10.28GKK323 pKa = 9.36PGSPSGAPARR333 pKa = 11.84SVHH336 pKa = 5.66EE337 pKa = 4.44PKK339 pKa = 10.69PMAATVRR346 pKa = 11.84SVVQQ350 pKa = 3.29
MM1 pKa = 7.48SLDD4 pKa = 3.4NQHH7 pKa = 6.07RR8 pKa = 11.84HH9 pKa = 4.71SAALEE14 pKa = 4.0CLPTARR20 pKa = 11.84KK21 pKa = 9.03RR22 pKa = 11.84AGIRR26 pKa = 11.84AHH28 pKa = 5.85LAVYY32 pKa = 10.09RR33 pKa = 11.84RR34 pKa = 11.84LIKK37 pKa = 10.3HH38 pKa = 6.52RR39 pKa = 11.84SLGDD43 pKa = 2.99IFKK46 pKa = 10.49FLSICSTRR54 pKa = 11.84EE55 pKa = 3.79ATEE58 pKa = 4.05DD59 pKa = 3.15AQFRR63 pKa = 11.84IFFEE67 pKa = 4.12VTLGKK72 pKa = 10.47RR73 pKa = 11.84IADD76 pKa = 3.44CVLTVEE82 pKa = 4.97SKK84 pKa = 10.16NQKK87 pKa = 6.26TCYY90 pKa = 9.65VVEE93 pKa = 4.86LKK95 pKa = 9.6TCLSGFVFPGSAIKK109 pKa = 10.43ISQRR113 pKa = 11.84RR114 pKa = 11.84QGLEE118 pKa = 3.57QLTDD122 pKa = 3.31SVAYY126 pKa = 9.51IGRR129 pKa = 11.84AAPKK133 pKa = 10.09GHH135 pKa = 6.51EE136 pKa = 4.15NWSVRR141 pKa = 11.84PLLLFKK147 pKa = 10.55NQKK150 pKa = 5.53TLKK153 pKa = 8.91TIHH156 pKa = 6.4TEE158 pKa = 3.89SSAFPPTFINTTSVALNGFFNQWEE182 pKa = 4.24DD183 pKa = 3.25ARR185 pKa = 11.84VRR187 pKa = 11.84KK188 pKa = 8.62MLYY191 pKa = 9.97AIPAKK196 pKa = 9.58THH198 pKa = 5.32ATNNLEE204 pKa = 4.21FLDD207 pKa = 4.34PASKK211 pKa = 10.09QRR213 pKa = 11.84SVYY216 pKa = 10.4SQTIVNGGKK225 pKa = 9.91KK226 pKa = 9.79KK227 pKa = 10.08RR228 pKa = 11.84VRR230 pKa = 11.84DD231 pKa = 3.75AKK233 pKa = 10.05STAGAQGSRR242 pKa = 11.84AEE244 pKa = 4.74KK245 pKa = 10.38KK246 pKa = 9.19PAPARR251 pKa = 11.84ARR253 pKa = 11.84QRR255 pKa = 11.84AANAPTGNGDD265 pKa = 3.35GYY267 pKa = 11.32ARR269 pKa = 11.84HH270 pKa = 6.62RR271 pKa = 11.84DD272 pKa = 3.69DD273 pKa = 4.01PRR275 pKa = 11.84HH276 pKa = 5.03GRR278 pKa = 11.84RR279 pKa = 11.84FASTEE284 pKa = 3.51QKK286 pKa = 9.74PRR288 pKa = 11.84RR289 pKa = 11.84NRR291 pKa = 11.84TVNRR295 pKa = 11.84PATQNRR301 pKa = 11.84PSDD304 pKa = 2.92AWRR307 pKa = 11.84HH308 pKa = 4.11VRR310 pKa = 11.84GHH312 pKa = 5.96NSPRR316 pKa = 11.84GRR318 pKa = 11.84GFYY321 pKa = 10.28GKK323 pKa = 9.36PGSPSGAPARR333 pKa = 11.84SVHH336 pKa = 5.66EE337 pKa = 4.44PKK339 pKa = 10.69PMAATVRR346 pKa = 11.84SVVQQ350 pKa = 3.29
Molecular weight: 39.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
41158 |
70 |
2554 |
433.2 |
48.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.889 ± 0.237 | 2.488 ± 0.151 |
4.84 ± 0.153 | 5.005 ± 0.133 |
4.39 ± 0.148 | 5.919 ± 0.194 |
2.707 ± 0.094 | 4.816 ± 0.162 |
4.087 ± 0.197 | 10.183 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.065 ± 0.106 | 4.019 ± 0.156 |
6.434 ± 0.284 | 3.705 ± 0.152 |
5.979 ± 0.194 | 7.389 ± 0.181 |
6.521 ± 0.252 | 7.199 ± 0.198 |
1.2 ± 0.083 | 3.166 ± 0.12 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
