
Abalone shriveling syndrome-associated virus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
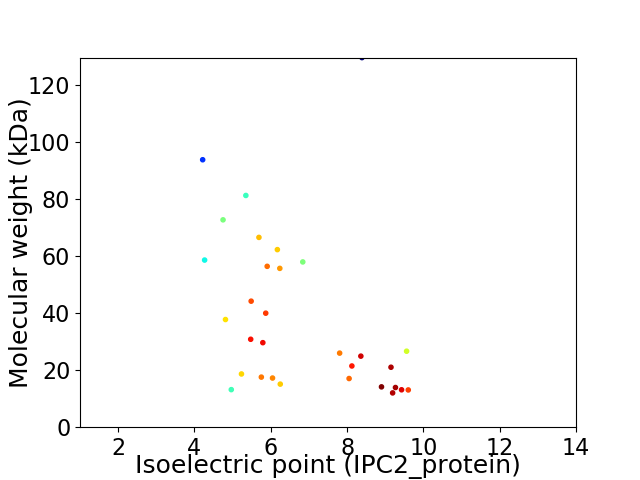
Virtual 2D-PAGE plot for 31 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
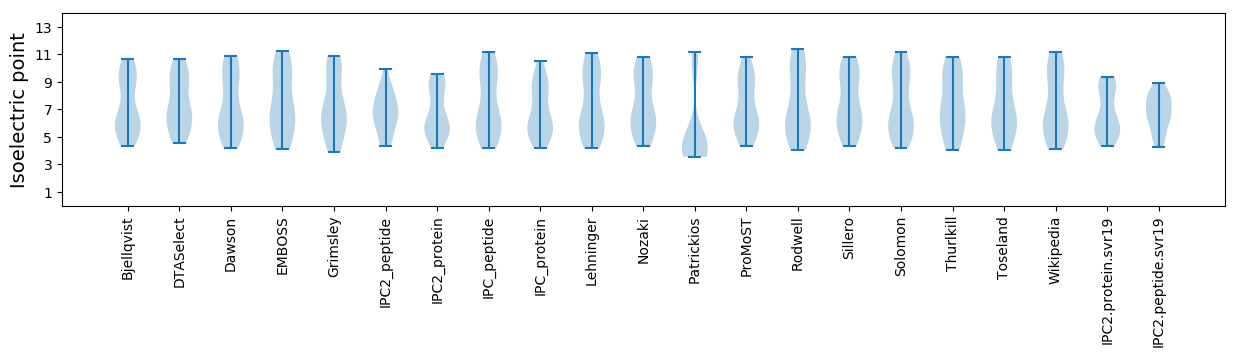
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8Q5C9|B8Q5C9_9CAUD Uncharacterized protein OS=Abalone shriveling syndrome-associated virus OX=491893 PE=4 SV=1
MM1 pKa = 8.38VEE3 pKa = 4.15FSKK6 pKa = 10.72IKK8 pKa = 10.46SDD10 pKa = 3.79LGSVAYY16 pKa = 9.79IEE18 pKa = 5.95DD19 pKa = 3.77SADD22 pKa = 3.56LNKK25 pKa = 9.89VPLSAVDD32 pKa = 3.38AGGFFYY38 pKa = 10.35WKK40 pKa = 9.71AANNMTPEE48 pKa = 4.66LSSADD53 pKa = 2.94IGKK56 pKa = 10.3AYY58 pKa = 10.45GITTVDD64 pKa = 2.88ASGAVTFGGVDD75 pKa = 3.42AFLLDD80 pKa = 4.42DD81 pKa = 4.08ATASIINQAGIIEE94 pKa = 4.94PPTLSITGTNTINVGPFKK112 pKa = 10.77AVFFDD117 pKa = 3.88QPAGAVGNVRR127 pKa = 11.84RR128 pKa = 11.84VVDD131 pKa = 3.89SPVQQTITINAVGSSNQDD149 pKa = 2.77VTYY152 pKa = 11.02LKK154 pKa = 10.16FLPSGSFAQEE164 pKa = 3.84TDD166 pKa = 3.48DD167 pKa = 3.87PTADD171 pKa = 5.26RR172 pKa = 11.84IIEE175 pKa = 4.11DD176 pKa = 3.61NYY178 pKa = 10.13IVRR181 pKa = 11.84ILHH184 pKa = 6.04PNGGKK189 pKa = 9.39IVGLSPVSTVYY200 pKa = 11.1SNPQQNLQQYY210 pKa = 8.18SSRR213 pKa = 11.84IGIDD217 pKa = 3.2GNDD220 pKa = 3.11ILFSISNNSLRR231 pKa = 11.84QTGANARR238 pKa = 11.84IFGYY242 pKa = 10.66NIADD246 pKa = 3.63GSGTGNRR253 pKa = 11.84SLRR256 pKa = 11.84TIAVEE261 pKa = 5.76DD262 pKa = 3.39ITADD266 pKa = 3.63YY267 pKa = 11.27YY268 pKa = 11.8SFDD271 pKa = 3.66STEE274 pKa = 3.65QTTFSDD280 pKa = 3.96LRR282 pKa = 11.84RR283 pKa = 11.84TYY285 pKa = 11.23KK286 pKa = 10.61LDD288 pKa = 3.86DD289 pKa = 4.18PAAGTSSALANFGIIAVYY307 pKa = 10.1ILASGSYY314 pKa = 10.64YY315 pKa = 10.93YY316 pKa = 10.87LAPQKK321 pKa = 10.65NYY323 pKa = 9.11STAALARR330 pKa = 11.84SDD332 pKa = 3.81LLFYY336 pKa = 10.48LTGTVVPDD344 pKa = 4.1FLSSSFAALLGAIVVPATVTDD365 pKa = 4.33ANISTVLTTTISNISLASSGGGVQTLTIDD394 pKa = 3.85PSTGTNDD401 pKa = 4.26DD402 pKa = 3.73VVGTNGTNFILRR414 pKa = 11.84KK415 pKa = 9.41EE416 pKa = 4.04NAFNVTGGNEE426 pKa = 3.66GDD428 pKa = 3.78VLVKK432 pKa = 10.48RR433 pKa = 11.84GTSFAVEE440 pKa = 4.7PIANQGVPRR449 pKa = 11.84FNLGTDD455 pKa = 2.32RR456 pKa = 11.84GGFVKK461 pKa = 10.73LDD463 pKa = 3.54VSNGAIVSSNGDD475 pKa = 3.16IASDD479 pKa = 4.13FSSAIIRR486 pKa = 11.84QVNDD490 pKa = 3.12AYY492 pKa = 10.89YY493 pKa = 10.03ISLISISTTKK503 pKa = 10.1IPVWISSYY511 pKa = 11.25VLNTDD516 pKa = 3.22GNMMPVTPTADD527 pKa = 2.9ISIDD531 pKa = 3.87FIVPVVLTPAQPEE544 pKa = 3.88FLLRR548 pKa = 11.84YY549 pKa = 9.62ILVNKK554 pKa = 9.95SS555 pKa = 2.69
MM1 pKa = 8.38VEE3 pKa = 4.15FSKK6 pKa = 10.72IKK8 pKa = 10.46SDD10 pKa = 3.79LGSVAYY16 pKa = 9.79IEE18 pKa = 5.95DD19 pKa = 3.77SADD22 pKa = 3.56LNKK25 pKa = 9.89VPLSAVDD32 pKa = 3.38AGGFFYY38 pKa = 10.35WKK40 pKa = 9.71AANNMTPEE48 pKa = 4.66LSSADD53 pKa = 2.94IGKK56 pKa = 10.3AYY58 pKa = 10.45GITTVDD64 pKa = 2.88ASGAVTFGGVDD75 pKa = 3.42AFLLDD80 pKa = 4.42DD81 pKa = 4.08ATASIINQAGIIEE94 pKa = 4.94PPTLSITGTNTINVGPFKK112 pKa = 10.77AVFFDD117 pKa = 3.88QPAGAVGNVRR127 pKa = 11.84RR128 pKa = 11.84VVDD131 pKa = 3.89SPVQQTITINAVGSSNQDD149 pKa = 2.77VTYY152 pKa = 11.02LKK154 pKa = 10.16FLPSGSFAQEE164 pKa = 3.84TDD166 pKa = 3.48DD167 pKa = 3.87PTADD171 pKa = 5.26RR172 pKa = 11.84IIEE175 pKa = 4.11DD176 pKa = 3.61NYY178 pKa = 10.13IVRR181 pKa = 11.84ILHH184 pKa = 6.04PNGGKK189 pKa = 9.39IVGLSPVSTVYY200 pKa = 11.1SNPQQNLQQYY210 pKa = 8.18SSRR213 pKa = 11.84IGIDD217 pKa = 3.2GNDD220 pKa = 3.11ILFSISNNSLRR231 pKa = 11.84QTGANARR238 pKa = 11.84IFGYY242 pKa = 10.66NIADD246 pKa = 3.63GSGTGNRR253 pKa = 11.84SLRR256 pKa = 11.84TIAVEE261 pKa = 5.76DD262 pKa = 3.39ITADD266 pKa = 3.63YY267 pKa = 11.27YY268 pKa = 11.8SFDD271 pKa = 3.66STEE274 pKa = 3.65QTTFSDD280 pKa = 3.96LRR282 pKa = 11.84RR283 pKa = 11.84TYY285 pKa = 11.23KK286 pKa = 10.61LDD288 pKa = 3.86DD289 pKa = 4.18PAAGTSSALANFGIIAVYY307 pKa = 10.1ILASGSYY314 pKa = 10.64YY315 pKa = 10.93YY316 pKa = 10.87LAPQKK321 pKa = 10.65NYY323 pKa = 9.11STAALARR330 pKa = 11.84SDD332 pKa = 3.81LLFYY336 pKa = 10.48LTGTVVPDD344 pKa = 4.1FLSSSFAALLGAIVVPATVTDD365 pKa = 4.33ANISTVLTTTISNISLASSGGGVQTLTIDD394 pKa = 3.85PSTGTNDD401 pKa = 4.26DD402 pKa = 3.73VVGTNGTNFILRR414 pKa = 11.84KK415 pKa = 9.41EE416 pKa = 4.04NAFNVTGGNEE426 pKa = 3.66GDD428 pKa = 3.78VLVKK432 pKa = 10.48RR433 pKa = 11.84GTSFAVEE440 pKa = 4.7PIANQGVPRR449 pKa = 11.84FNLGTDD455 pKa = 2.32RR456 pKa = 11.84GGFVKK461 pKa = 10.73LDD463 pKa = 3.54VSNGAIVSSNGDD475 pKa = 3.16IASDD479 pKa = 4.13FSSAIIRR486 pKa = 11.84QVNDD490 pKa = 3.12AYY492 pKa = 10.89YY493 pKa = 10.03ISLISISTTKK503 pKa = 10.1IPVWISSYY511 pKa = 11.25VLNTDD516 pKa = 3.22GNMMPVTPTADD527 pKa = 2.9ISIDD531 pKa = 3.87FIVPVVLTPAQPEE544 pKa = 3.88FLLRR548 pKa = 11.84YY549 pKa = 9.62ILVNKK554 pKa = 9.95SS555 pKa = 2.69
Molecular weight: 58.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8Q5C2|B8Q5C2_9CAUD Uncharacterized protein OS=Abalone shriveling syndrome-associated virus OX=491893 PE=4 SV=1
MM1 pKa = 7.58ALFAFAALASIAAAGFGACRR21 pKa = 11.84NSRR24 pKa = 11.84EE25 pKa = 3.67NSRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.64VLLRR34 pKa = 11.84NEE36 pKa = 4.13LVAQEE41 pKa = 4.13EE42 pKa = 4.35LLNRR46 pKa = 11.84EE47 pKa = 3.75RR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84AAGINAFQRR60 pKa = 11.84RR61 pKa = 11.84GSTAFDD67 pKa = 4.2DD68 pKa = 5.2LDD70 pKa = 5.55DD71 pKa = 4.41EE72 pKa = 5.04FSLRR76 pKa = 11.84GNSLNANTYY85 pKa = 7.51SRR87 pKa = 11.84RR88 pKa = 11.84VANPGLTDD96 pKa = 3.58SFADD100 pKa = 4.12LSRR103 pKa = 11.84GVVKK107 pKa = 10.46AAEE110 pKa = 3.88FLINRR115 pKa = 11.84SKK117 pKa = 11.27LGGG120 pKa = 3.47
MM1 pKa = 7.58ALFAFAALASIAAAGFGACRR21 pKa = 11.84NSRR24 pKa = 11.84EE25 pKa = 3.67NSRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.64VLLRR34 pKa = 11.84NEE36 pKa = 4.13LVAQEE41 pKa = 4.13EE42 pKa = 4.35LLNRR46 pKa = 11.84EE47 pKa = 3.75RR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84AAGINAFQRR60 pKa = 11.84RR61 pKa = 11.84GSTAFDD67 pKa = 4.2DD68 pKa = 5.2LDD70 pKa = 5.55DD71 pKa = 4.41EE72 pKa = 5.04FSLRR76 pKa = 11.84GNSLNANTYY85 pKa = 7.51SRR87 pKa = 11.84RR88 pKa = 11.84VANPGLTDD96 pKa = 3.58SFADD100 pKa = 4.12LSRR103 pKa = 11.84GVVKK107 pKa = 10.46AAEE110 pKa = 3.88FLINRR115 pKa = 11.84SKK117 pKa = 11.27LGGG120 pKa = 3.47
Molecular weight: 13.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10892 |
115 |
1194 |
351.4 |
38.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.456 ± 0.547 | 0.826 ± 0.21 |
5.812 ± 0.356 | 4.912 ± 0.349 |
5.224 ± 0.252 | 6.023 ± 0.43 |
1.451 ± 0.243 | 6.721 ± 0.285 |
5.922 ± 0.418 | 9.236 ± 0.341 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.836 ± 0.168 | 5.913 ± 0.303 |
3.819 ± 0.248 | 3.452 ± 0.183 |
4.361 ± 0.308 | 8.336 ± 0.347 |
6.941 ± 0.365 | 6.62 ± 0.346 |
0.633 ± 0.089 | 3.507 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
