
San Bernardo virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
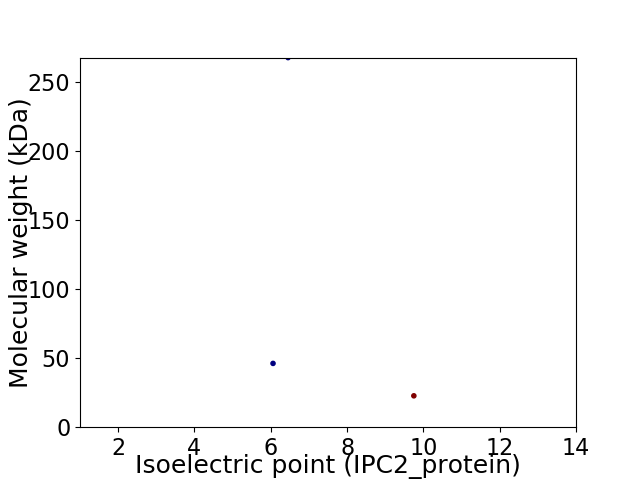
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7EIV6|A0A1U7EIV6_9VIRU Non-structural protein 3 OS=San Bernardo virus OX=1955199 PE=4 SV=1
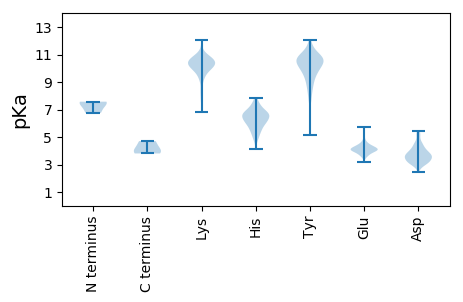
MM1 pKa = 7.49KK2 pKa = 10.36FLVLFVSVVNAAVFVKK18 pKa = 10.62DD19 pKa = 3.4VTRR22 pKa = 11.84QLATTRR28 pKa = 11.84EE29 pKa = 4.05RR30 pKa = 11.84YY31 pKa = 9.48ALYY34 pKa = 10.67QNLSPTLVKK43 pKa = 10.24IGNYY47 pKa = 9.22QISPFHH53 pKa = 7.35PDD55 pKa = 3.35PACVSVYY62 pKa = 8.95RR63 pKa = 11.84TNDD66 pKa = 2.48WFFAGCEE73 pKa = 4.21LPSHH77 pKa = 6.53CLGITVSVVEE87 pKa = 4.49KK88 pKa = 10.14KK89 pKa = 9.31WYY91 pKa = 8.56GQEE94 pKa = 4.06TVFCHH99 pKa = 5.5ATYY102 pKa = 10.53HH103 pKa = 6.76PDD105 pKa = 3.12RR106 pKa = 11.84TEE108 pKa = 3.54TYY110 pKa = 8.73EE111 pKa = 3.96FYY113 pKa = 10.86NIEE116 pKa = 4.03FRR118 pKa = 11.84PSEE121 pKa = 3.9RR122 pKa = 11.84KK123 pKa = 9.37VRR125 pKa = 11.84PKK127 pKa = 10.93SPLKK131 pKa = 10.81LYY133 pKa = 10.68GKK135 pKa = 10.53VITEE139 pKa = 4.11ISNLYY144 pKa = 8.62PVSFFEE150 pKa = 4.42YY151 pKa = 9.84FVVARR156 pKa = 11.84NEE158 pKa = 4.29SGFGLVPKK166 pKa = 10.46FCMEE170 pKa = 4.0KK171 pKa = 10.49LRR173 pKa = 11.84NGTQLPKK180 pKa = 10.5NVQLRR185 pKa = 11.84HH186 pKa = 5.5SEE188 pKa = 3.88GMVCVDD194 pKa = 3.01EE195 pKa = 4.71VNYY198 pKa = 10.51DD199 pKa = 3.3EE200 pKa = 5.74HH201 pKa = 7.47DD202 pKa = 3.49CHH204 pKa = 7.75PYY206 pKa = 9.35MFSTVMEE213 pKa = 4.55MIMFYY218 pKa = 10.73DD219 pKa = 4.4FPIEE223 pKa = 4.0FTDD226 pKa = 3.72VPFSQGSFAATNNKK240 pKa = 9.73AMGADD245 pKa = 3.21AYY247 pKa = 10.15VANHH251 pKa = 6.19ASEE254 pKa = 4.69VPSHH258 pKa = 6.33VFDD261 pKa = 4.87KK262 pKa = 10.55IGRR265 pKa = 11.84SFMVSFVKK273 pKa = 8.27MTSSDD278 pKa = 3.17PFFIVKK284 pKa = 8.01NTRR287 pKa = 11.84KK288 pKa = 10.27LEE290 pKa = 4.18VIQTHH295 pKa = 6.8CYY297 pKa = 10.41DD298 pKa = 3.59FHH300 pKa = 6.18THH302 pKa = 5.4YY303 pKa = 10.73KK304 pKa = 10.46SPFSNFLSTVSKK316 pKa = 10.1FIQDD320 pKa = 3.55EE321 pKa = 4.02LSEE324 pKa = 4.18FLRR327 pKa = 11.84FLKK330 pKa = 10.61KK331 pKa = 10.18FSKK334 pKa = 10.57SIAEE338 pKa = 3.69ILLFVVSEE346 pKa = 4.25LLLMVADD353 pKa = 5.42LIPYY357 pKa = 8.67TGEE360 pKa = 3.97FYY362 pKa = 10.45TGAFITLVFYY372 pKa = 9.15YY373 pKa = 7.78TTFNIGVSAVAGLLVYY389 pKa = 8.17FFRR392 pKa = 11.84IYY394 pKa = 10.19IDD396 pKa = 3.33SLIFF400 pKa = 4.02
MM1 pKa = 7.49KK2 pKa = 10.36FLVLFVSVVNAAVFVKK18 pKa = 10.62DD19 pKa = 3.4VTRR22 pKa = 11.84QLATTRR28 pKa = 11.84EE29 pKa = 4.05RR30 pKa = 11.84YY31 pKa = 9.48ALYY34 pKa = 10.67QNLSPTLVKK43 pKa = 10.24IGNYY47 pKa = 9.22QISPFHH53 pKa = 7.35PDD55 pKa = 3.35PACVSVYY62 pKa = 8.95RR63 pKa = 11.84TNDD66 pKa = 2.48WFFAGCEE73 pKa = 4.21LPSHH77 pKa = 6.53CLGITVSVVEE87 pKa = 4.49KK88 pKa = 10.14KK89 pKa = 9.31WYY91 pKa = 8.56GQEE94 pKa = 4.06TVFCHH99 pKa = 5.5ATYY102 pKa = 10.53HH103 pKa = 6.76PDD105 pKa = 3.12RR106 pKa = 11.84TEE108 pKa = 3.54TYY110 pKa = 8.73EE111 pKa = 3.96FYY113 pKa = 10.86NIEE116 pKa = 4.03FRR118 pKa = 11.84PSEE121 pKa = 3.9RR122 pKa = 11.84KK123 pKa = 9.37VRR125 pKa = 11.84PKK127 pKa = 10.93SPLKK131 pKa = 10.81LYY133 pKa = 10.68GKK135 pKa = 10.53VITEE139 pKa = 4.11ISNLYY144 pKa = 8.62PVSFFEE150 pKa = 4.42YY151 pKa = 9.84FVVARR156 pKa = 11.84NEE158 pKa = 4.29SGFGLVPKK166 pKa = 10.46FCMEE170 pKa = 4.0KK171 pKa = 10.49LRR173 pKa = 11.84NGTQLPKK180 pKa = 10.5NVQLRR185 pKa = 11.84HH186 pKa = 5.5SEE188 pKa = 3.88GMVCVDD194 pKa = 3.01EE195 pKa = 4.71VNYY198 pKa = 10.51DD199 pKa = 3.3EE200 pKa = 5.74HH201 pKa = 7.47DD202 pKa = 3.49CHH204 pKa = 7.75PYY206 pKa = 9.35MFSTVMEE213 pKa = 4.55MIMFYY218 pKa = 10.73DD219 pKa = 4.4FPIEE223 pKa = 4.0FTDD226 pKa = 3.72VPFSQGSFAATNNKK240 pKa = 9.73AMGADD245 pKa = 3.21AYY247 pKa = 10.15VANHH251 pKa = 6.19ASEE254 pKa = 4.69VPSHH258 pKa = 6.33VFDD261 pKa = 4.87KK262 pKa = 10.55IGRR265 pKa = 11.84SFMVSFVKK273 pKa = 8.27MTSSDD278 pKa = 3.17PFFIVKK284 pKa = 8.01NTRR287 pKa = 11.84KK288 pKa = 10.27LEE290 pKa = 4.18VIQTHH295 pKa = 6.8CYY297 pKa = 10.41DD298 pKa = 3.59FHH300 pKa = 6.18THH302 pKa = 5.4YY303 pKa = 10.73KK304 pKa = 10.46SPFSNFLSTVSKK316 pKa = 10.1FIQDD320 pKa = 3.55EE321 pKa = 4.02LSEE324 pKa = 4.18FLRR327 pKa = 11.84FLKK330 pKa = 10.61KK331 pKa = 10.18FSKK334 pKa = 10.57SIAEE338 pKa = 3.69ILLFVVSEE346 pKa = 4.25LLLMVADD353 pKa = 5.42LIPYY357 pKa = 8.67TGEE360 pKa = 3.97FYY362 pKa = 10.45TGAFITLVFYY372 pKa = 9.15YY373 pKa = 7.78TTFNIGVSAVAGLLVYY389 pKa = 8.17FFRR392 pKa = 11.84IYY394 pKa = 10.19IDD396 pKa = 3.33SLIFF400 pKa = 4.02
Molecular weight: 46.16 kDa
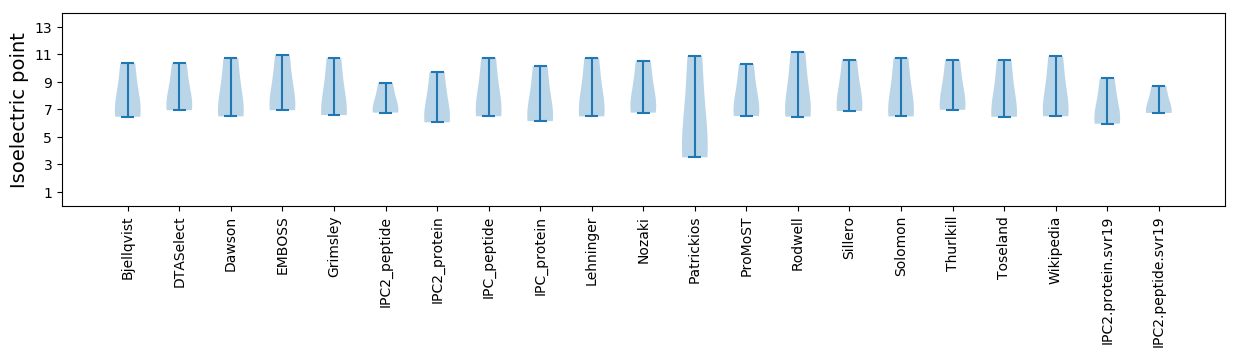
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7EJ03|A0A1U7EJ03_9VIRU Uncharacterized protein OS=San Bernardo virus OX=1955199 PE=4 SV=1
MM1 pKa = 6.76STNRR5 pKa = 11.84NTNKK9 pKa = 8.47NTKK12 pKa = 8.39QSSGARR18 pKa = 11.84VSAARR23 pKa = 11.84NTTLTKK29 pKa = 10.56AVVRR33 pKa = 11.84NAPKK37 pKa = 9.04RR38 pKa = 11.84TEE40 pKa = 3.95VSSFDD45 pKa = 3.8LNKK48 pKa = 9.99IVSDD52 pKa = 3.83VSSTFLNALNRR63 pKa = 11.84PMVLLSLVMVVALVFTHH80 pKa = 6.66QSDD83 pKa = 4.44FSSGAVGKK91 pKa = 10.0YY92 pKa = 8.94VADD95 pKa = 4.03RR96 pKa = 11.84AEE98 pKa = 4.33TNSLAKK104 pKa = 9.15WVHH107 pKa = 5.23EE108 pKa = 4.17NQTKK112 pKa = 10.21FLGLAIFTPAVLNSPEE128 pKa = 4.39KK129 pKa = 9.38IRR131 pKa = 11.84VVIGLATLLWVMLVPQASVVEE152 pKa = 4.25YY153 pKa = 10.73VLQALALHH161 pKa = 6.63SYY163 pKa = 10.29FRR165 pKa = 11.84VKK167 pKa = 10.26LQHH170 pKa = 5.98SRR172 pKa = 11.84LFIMAMVVVLYY183 pKa = 9.55FMGYY187 pKa = 7.75LTLVKK192 pKa = 10.54SPGSAAGSVNKK203 pKa = 10.17TNDD206 pKa = 2.84RR207 pKa = 3.81
MM1 pKa = 6.76STNRR5 pKa = 11.84NTNKK9 pKa = 8.47NTKK12 pKa = 8.39QSSGARR18 pKa = 11.84VSAARR23 pKa = 11.84NTTLTKK29 pKa = 10.56AVVRR33 pKa = 11.84NAPKK37 pKa = 9.04RR38 pKa = 11.84TEE40 pKa = 3.95VSSFDD45 pKa = 3.8LNKK48 pKa = 9.99IVSDD52 pKa = 3.83VSSTFLNALNRR63 pKa = 11.84PMVLLSLVMVVALVFTHH80 pKa = 6.66QSDD83 pKa = 4.44FSSGAVGKK91 pKa = 10.0YY92 pKa = 8.94VADD95 pKa = 4.03RR96 pKa = 11.84AEE98 pKa = 4.33TNSLAKK104 pKa = 9.15WVHH107 pKa = 5.23EE108 pKa = 4.17NQTKK112 pKa = 10.21FLGLAIFTPAVLNSPEE128 pKa = 4.39KK129 pKa = 9.38IRR131 pKa = 11.84VVIGLATLLWVMLVPQASVVEE152 pKa = 4.25YY153 pKa = 10.73VLQALALHH161 pKa = 6.63SYY163 pKa = 10.29FRR165 pKa = 11.84VKK167 pKa = 10.26LQHH170 pKa = 5.98SRR172 pKa = 11.84LFIMAMVVVLYY183 pKa = 9.55FMGYY187 pKa = 7.75LTLVKK192 pKa = 10.54SPGSAAGSVNKK203 pKa = 10.17TNDD206 pKa = 2.84RR207 pKa = 3.81
Molecular weight: 22.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2944 |
207 |
2337 |
981.3 |
112.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.537 ± 0.86 | 2.14 ± 0.515 |
6.046 ± 1.358 | 5.876 ± 0.768 |
6.352 ± 1.152 | 4.178 ± 0.118 |
2.582 ± 0.149 | 4.993 ± 0.587 |
6.352 ± 0.386 | 8.798 ± 0.57 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.717 ± 0.158 | 5.197 ± 0.397 |
3.668 ± 0.337 | 2.48 ± 0.089 |
5.74 ± 0.77 | 8.22 ± 0.383 |
5.673 ± 0.497 | 8.865 ± 1.383 |
0.543 ± 0.09 | 4.042 ± 0.534 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
