
Passion fruit mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
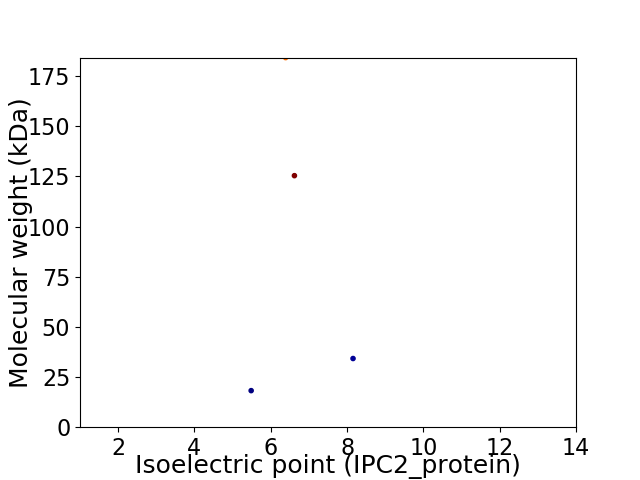
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
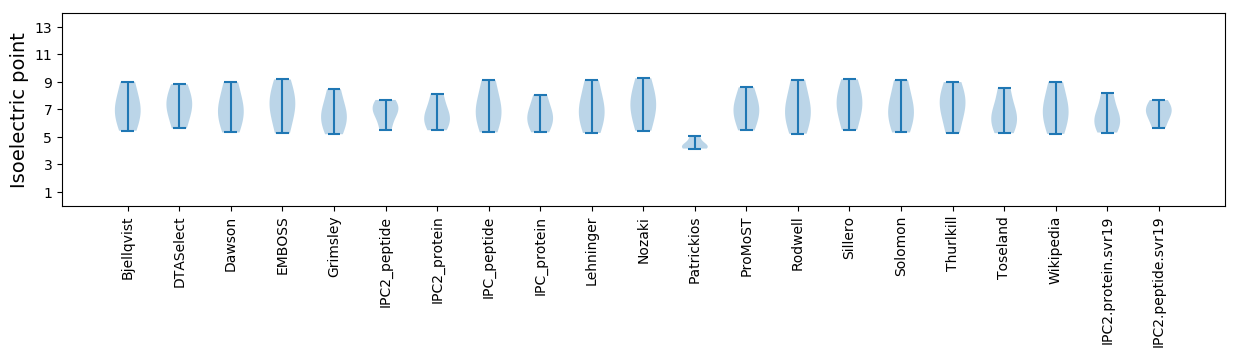
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6KJM7|F6KJM7_9VIRU Capsid protein OS=Passion fruit mosaic virus OX=1032457 PE=3 SV=1
MM1 pKa = 8.08PYY3 pKa = 10.29QPVSLQTLPWLSANWADD20 pKa = 3.73YY21 pKa = 7.9KK22 pKa = 10.95TLLSVLRR29 pKa = 11.84ATSATSFQTQAGRR42 pKa = 11.84DD43 pKa = 3.81SIRR46 pKa = 11.84SQLAGCVSEE55 pKa = 4.54LVQVNVRR62 pKa = 11.84FPEE65 pKa = 4.59RR66 pKa = 11.84YY67 pKa = 8.8LVYY70 pKa = 11.03VNDD73 pKa = 4.45PSISDD78 pKa = 3.11VWGALLKK85 pKa = 10.38ATDD88 pKa = 3.55TKK90 pKa = 11.41NRR92 pKa = 11.84IIEE95 pKa = 4.09VDD97 pKa = 3.39NEE99 pKa = 4.15RR100 pKa = 11.84NPSNNEE106 pKa = 3.4IEE108 pKa = 4.62SVTRR112 pKa = 11.84RR113 pKa = 11.84VDD115 pKa = 3.43DD116 pKa = 4.03ASVAIRR122 pKa = 11.84INVEE126 pKa = 3.79RR127 pKa = 11.84LLKK130 pKa = 10.87LLGEE134 pKa = 4.14VHH136 pKa = 6.33GVYY139 pKa = 10.48DD140 pKa = 3.44RR141 pKa = 11.84ALFEE145 pKa = 4.32QVSGLRR151 pKa = 11.84WADD154 pKa = 3.56DD155 pKa = 3.59SAPASTSKK163 pKa = 11.13
MM1 pKa = 8.08PYY3 pKa = 10.29QPVSLQTLPWLSANWADD20 pKa = 3.73YY21 pKa = 7.9KK22 pKa = 10.95TLLSVLRR29 pKa = 11.84ATSATSFQTQAGRR42 pKa = 11.84DD43 pKa = 3.81SIRR46 pKa = 11.84SQLAGCVSEE55 pKa = 4.54LVQVNVRR62 pKa = 11.84FPEE65 pKa = 4.59RR66 pKa = 11.84YY67 pKa = 8.8LVYY70 pKa = 11.03VNDD73 pKa = 4.45PSISDD78 pKa = 3.11VWGALLKK85 pKa = 10.38ATDD88 pKa = 3.55TKK90 pKa = 11.41NRR92 pKa = 11.84IIEE95 pKa = 4.09VDD97 pKa = 3.39NEE99 pKa = 4.15RR100 pKa = 11.84NPSNNEE106 pKa = 3.4IEE108 pKa = 4.62SVTRR112 pKa = 11.84RR113 pKa = 11.84VDD115 pKa = 3.43DD116 pKa = 4.03ASVAIRR122 pKa = 11.84INVEE126 pKa = 3.79RR127 pKa = 11.84LLKK130 pKa = 10.87LLGEE134 pKa = 4.14VHH136 pKa = 6.33GVYY139 pKa = 10.48DD140 pKa = 3.44RR141 pKa = 11.84ALFEE145 pKa = 4.32QVSGLRR151 pKa = 11.84WADD154 pKa = 3.56DD155 pKa = 3.59SAPASTSKK163 pKa = 11.13
Molecular weight: 18.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6KJM7|F6KJM7_9VIRU Capsid protein OS=Passion fruit mosaic virus OX=1032457 PE=3 SV=1
MM1 pKa = 7.74SLRR4 pKa = 11.84AQNAQDD10 pKa = 3.65VVPPLDD16 pKa = 3.58YY17 pKa = 11.12LNPSNFVKK25 pKa = 9.62TQLLGLEE32 pKa = 3.98RR33 pKa = 11.84LGLVKK38 pKa = 10.48SPKK41 pKa = 10.04KK42 pKa = 9.67MCALMEE48 pKa = 4.21TDD50 pKa = 3.63VVKK53 pKa = 10.72ISPRR57 pKa = 11.84EE58 pKa = 3.68RR59 pKa = 11.84TVFPLRR65 pKa = 11.84LTEE68 pKa = 4.48KK69 pKa = 10.51IKK71 pKa = 11.05DD72 pKa = 3.55LGNYY76 pKa = 8.82KK77 pKa = 10.45YY78 pKa = 11.55VMFLACVVSGRR89 pKa = 11.84WHH91 pKa = 6.05VPTTVKK97 pKa = 11.1GEE99 pKa = 4.21VFLSLMDD106 pKa = 3.53RR107 pKa = 11.84RR108 pKa = 11.84LTNEE112 pKa = 3.89RR113 pKa = 11.84EE114 pKa = 4.44SVILTANAKK123 pKa = 9.76PSVSEE128 pKa = 3.7FQIRR132 pKa = 11.84IHH134 pKa = 6.42PRR136 pKa = 11.84YY137 pKa = 10.88SMVSSDD143 pKa = 4.71AMQEE147 pKa = 3.88PLEE150 pKa = 4.39LFVHH154 pKa = 6.08VKK156 pKa = 9.16GLQMAEE162 pKa = 4.11GFSPLSLEE170 pKa = 3.7IAFCVVCCDD179 pKa = 3.57VVIAKK184 pKa = 9.15SLKK187 pKa = 9.78MKK189 pKa = 10.34ILQNTDD195 pKa = 2.35KK196 pKa = 10.88FGNAEE201 pKa = 4.11GEE203 pKa = 4.43VGSEE207 pKa = 4.16GLDD210 pKa = 3.56DD211 pKa = 5.37LLSGSNMPSIRR222 pKa = 11.84TSAFNSRR229 pKa = 11.84RR230 pKa = 11.84SVNANPKK237 pKa = 10.06RR238 pKa = 11.84VFKK241 pKa = 10.75GKK243 pKa = 10.07KK244 pKa = 7.95GNVWSEE250 pKa = 3.89AKK252 pKa = 10.58DD253 pKa = 3.4FAGVRR258 pKa = 11.84RR259 pKa = 11.84KK260 pKa = 10.36KK261 pKa = 10.9GVVEE265 pKa = 4.21SDD267 pKa = 3.12MLTSNAEE274 pKa = 4.28STVSNYY280 pKa = 9.28STDD283 pKa = 3.34QSDD286 pKa = 3.72ALSTGFSTDD295 pKa = 2.05TSMAVGQLGRR305 pKa = 11.84LQDD308 pKa = 3.58TSS310 pKa = 3.31
MM1 pKa = 7.74SLRR4 pKa = 11.84AQNAQDD10 pKa = 3.65VVPPLDD16 pKa = 3.58YY17 pKa = 11.12LNPSNFVKK25 pKa = 9.62TQLLGLEE32 pKa = 3.98RR33 pKa = 11.84LGLVKK38 pKa = 10.48SPKK41 pKa = 10.04KK42 pKa = 9.67MCALMEE48 pKa = 4.21TDD50 pKa = 3.63VVKK53 pKa = 10.72ISPRR57 pKa = 11.84EE58 pKa = 3.68RR59 pKa = 11.84TVFPLRR65 pKa = 11.84LTEE68 pKa = 4.48KK69 pKa = 10.51IKK71 pKa = 11.05DD72 pKa = 3.55LGNYY76 pKa = 8.82KK77 pKa = 10.45YY78 pKa = 11.55VMFLACVVSGRR89 pKa = 11.84WHH91 pKa = 6.05VPTTVKK97 pKa = 11.1GEE99 pKa = 4.21VFLSLMDD106 pKa = 3.53RR107 pKa = 11.84RR108 pKa = 11.84LTNEE112 pKa = 3.89RR113 pKa = 11.84EE114 pKa = 4.44SVILTANAKK123 pKa = 9.76PSVSEE128 pKa = 3.7FQIRR132 pKa = 11.84IHH134 pKa = 6.42PRR136 pKa = 11.84YY137 pKa = 10.88SMVSSDD143 pKa = 4.71AMQEE147 pKa = 3.88PLEE150 pKa = 4.39LFVHH154 pKa = 6.08VKK156 pKa = 9.16GLQMAEE162 pKa = 4.11GFSPLSLEE170 pKa = 3.7IAFCVVCCDD179 pKa = 3.57VVIAKK184 pKa = 9.15SLKK187 pKa = 9.78MKK189 pKa = 10.34ILQNTDD195 pKa = 2.35KK196 pKa = 10.88FGNAEE201 pKa = 4.11GEE203 pKa = 4.43VGSEE207 pKa = 4.16GLDD210 pKa = 3.56DD211 pKa = 5.37LLSGSNMPSIRR222 pKa = 11.84TSAFNSRR229 pKa = 11.84RR230 pKa = 11.84SVNANPKK237 pKa = 10.06RR238 pKa = 11.84VFKK241 pKa = 10.75GKK243 pKa = 10.07KK244 pKa = 7.95GNVWSEE250 pKa = 3.89AKK252 pKa = 10.58DD253 pKa = 3.4FAGVRR258 pKa = 11.84RR259 pKa = 11.84KK260 pKa = 10.36KK261 pKa = 10.9GVVEE265 pKa = 4.21SDD267 pKa = 3.12MLTSNAEE274 pKa = 4.28STVSNYY280 pKa = 9.28STDD283 pKa = 3.34QSDD286 pKa = 3.72ALSTGFSTDD295 pKa = 2.05TSMAVGQLGRR305 pKa = 11.84LQDD308 pKa = 3.58TSS310 pKa = 3.31
Molecular weight: 34.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3184 |
163 |
1611 |
796.0 |
90.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.151 ± 0.538 | 2.041 ± 0.254 |
6.344 ± 0.255 | 5.842 ± 0.16 |
5.119 ± 0.53 | 4.931 ± 0.25 |
1.696 ± 0.223 | 4.648 ± 0.407 |
6.376 ± 0.469 | 9.108 ± 0.27 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.326 | 4.68 ± 0.165 |
3.832 ± 0.102 | 3.455 ± 0.131 |
5.842 ± 0.365 | 7.538 ± 0.772 |
6.91 ± 0.411 | 9.265 ± 0.37 |
1.225 ± 0.178 | 3.612 ± 0.418 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
