
Desulfonema ishimotonii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfonema
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
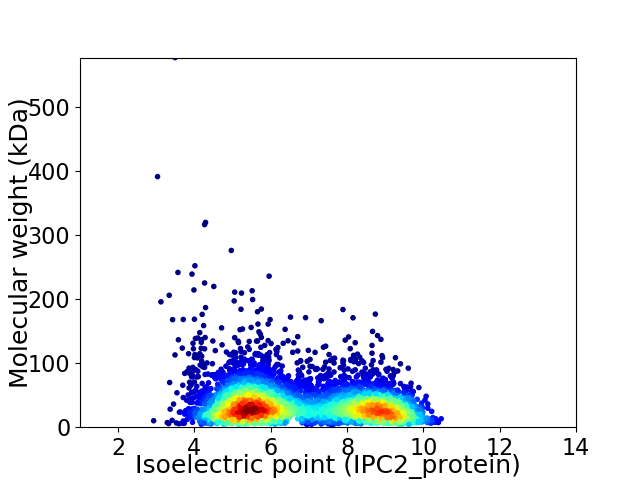
Virtual 2D-PAGE plot for 5012 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A401FWM4|A0A401FWM4_9DELT ABC transporter ATP-binding protein OS=Desulfonema ishimotonii OX=45657 GN=DENIS_2337 PE=4 SV=1
MM1 pKa = 7.49TSGEE5 pKa = 4.48YY6 pKa = 8.87FTVRR10 pKa = 11.84YY11 pKa = 9.28EE12 pKa = 4.07SGGVKK17 pKa = 10.16LVVSEE22 pKa = 4.71VISEE26 pKa = 3.86FSIGLTISPSGSGRR40 pKa = 11.84VLIEE44 pKa = 3.86EE45 pKa = 4.52EE46 pKa = 4.46GDD48 pKa = 3.8GEE50 pKa = 4.53TGTTGLTFTCPDD62 pKa = 3.76DD63 pKa = 5.52CVDD66 pKa = 5.77SDD68 pKa = 4.34TEE70 pKa = 4.43CNSGFCKK77 pKa = 10.36KK78 pKa = 10.17YY79 pKa = 9.06FAPGARR85 pKa = 11.84LKK87 pKa = 10.0LTATPGSGYY96 pKa = 10.75LFDD99 pKa = 3.57TWDD102 pKa = 3.98GEE104 pKa = 4.66VEE106 pKa = 4.32STSDD110 pKa = 3.41PKK112 pKa = 10.61IVYY115 pKa = 10.22VDD117 pKa = 3.68MAADD121 pKa = 3.4QSVTARR127 pKa = 11.84FEE129 pKa = 3.96ISQPLPDD136 pKa = 4.6RR137 pKa = 11.84DD138 pKa = 3.9GDD140 pKa = 4.15GTPDD144 pKa = 3.69TQDD147 pKa = 3.22GCPDD151 pKa = 3.98DD152 pKa = 4.16PRR154 pKa = 11.84KK155 pKa = 8.59TSPGACGCGVADD167 pKa = 4.16TDD169 pKa = 4.27SDD171 pKa = 3.91GDD173 pKa = 3.87EE174 pKa = 4.57TPDD177 pKa = 5.25CNDD180 pKa = 3.7EE181 pKa = 4.98CPADD185 pKa = 3.99PNKK188 pKa = 9.52TAPGQCGCGVADD200 pKa = 4.02TDD202 pKa = 4.27SDD204 pKa = 3.91GDD206 pKa = 3.87EE207 pKa = 4.57TPDD210 pKa = 5.29CNDD213 pKa = 3.9EE214 pKa = 5.15CPDD217 pKa = 4.71DD218 pKa = 4.48PDD220 pKa = 3.64KK221 pKa = 11.16TSPGACGCGVADD233 pKa = 4.24TDD235 pKa = 4.16SDD237 pKa = 3.8GDD239 pKa = 3.88ATPDD243 pKa = 3.93CNDD246 pKa = 3.99RR247 pKa = 11.84CPDD250 pKa = 4.29DD251 pKa = 4.24PDD253 pKa = 3.42KK254 pKa = 11.38TEE256 pKa = 5.01PGQCGCGLSDD266 pKa = 4.99ADD268 pKa = 3.92SDD270 pKa = 4.09GDD272 pKa = 4.02GFSDD276 pKa = 5.87CRR278 pKa = 11.84DD279 pKa = 3.41QCPDD283 pKa = 4.01DD284 pKa = 4.49PGKK287 pKa = 8.32TLPGQCGCGVADD299 pKa = 4.43TDD301 pKa = 3.74TDD303 pKa = 3.93GDD305 pKa = 4.01GTADD309 pKa = 4.9CNDD312 pKa = 4.02KK313 pKa = 11.53CPDD316 pKa = 4.16DD317 pKa = 4.26PNKK320 pKa = 9.94TFPGQCGCGVVDD332 pKa = 4.29TDD334 pKa = 4.36SDD336 pKa = 4.18GDD338 pKa = 3.93GTPDD342 pKa = 5.32CFDD345 pKa = 4.08KK346 pKa = 11.3CPNDD350 pKa = 3.66ADD352 pKa = 3.77KK353 pKa = 11.11TEE355 pKa = 4.89PGQCGCGVADD365 pKa = 4.99ADD367 pKa = 4.32SDD369 pKa = 4.29GDD371 pKa = 4.05GTPDD375 pKa = 5.26CSDD378 pKa = 3.76KK379 pKa = 11.35CPNDD383 pKa = 3.6PDD385 pKa = 3.59KK386 pKa = 11.31TEE388 pKa = 4.91PGQCGCGIADD398 pKa = 3.99TDD400 pKa = 3.85SDD402 pKa = 4.16GDD404 pKa = 4.02GVADD408 pKa = 4.17CEE410 pKa = 4.47SEE412 pKa = 4.29AVSGIPVPISPEE424 pKa = 3.69NGEE427 pKa = 4.32TVAEE431 pKa = 4.29DD432 pKa = 3.58SVTLVAQVDD441 pKa = 3.84LGSTVDD447 pKa = 4.58GPMEE451 pKa = 4.37IYY453 pKa = 8.94WWWRR457 pKa = 11.84PVDD460 pKa = 3.84KK461 pKa = 10.74KK462 pKa = 10.62CTVSSEE468 pKa = 3.98TTEE471 pKa = 4.08AYY473 pKa = 10.01RR474 pKa = 11.84GLAEE478 pKa = 4.58LRR480 pKa = 11.84ISGLRR485 pKa = 11.84DD486 pKa = 2.97GLQYY490 pKa = 11.13AWQVGVRR497 pKa = 11.84NPASGEE503 pKa = 3.8IFISDD508 pKa = 3.32EE509 pKa = 4.07VFFTVGASEE518 pKa = 4.42VEE520 pKa = 4.23VPFRR524 pKa = 11.84VDD526 pKa = 4.66AGTTLNDD533 pKa = 2.91YY534 pKa = 11.18RR535 pKa = 11.84MYY537 pKa = 10.98SFSLWPEE544 pKa = 4.04DD545 pKa = 4.45ADD547 pKa = 4.71AADD550 pKa = 4.27LLGDD554 pKa = 3.85AVGEE558 pKa = 4.03YY559 pKa = 8.24DD560 pKa = 3.72TKK562 pKa = 10.68FFKK565 pKa = 10.51IGTYY569 pKa = 10.36DD570 pKa = 3.67PEE572 pKa = 3.73QSRR575 pKa = 11.84YY576 pKa = 8.54VEE578 pKa = 4.15YY579 pKa = 10.72DD580 pKa = 2.7SGAITIVPGQAYY592 pKa = 7.88WFLARR597 pKa = 11.84NGLEE601 pKa = 3.68ASIRR605 pKa = 11.84GIKK608 pKa = 10.27VSVAEE613 pKa = 4.64NIHH616 pKa = 4.29VHH618 pKa = 5.8LRR620 pKa = 11.84YY621 pKa = 10.36DD622 pKa = 3.43EE623 pKa = 4.14TRR625 pKa = 11.84EE626 pKa = 4.15KK627 pKa = 9.22GWNMIAPPNDD637 pKa = 2.92GDD639 pKa = 4.03YY640 pKa = 10.67FRR642 pKa = 11.84KK643 pKa = 10.21DD644 pKa = 3.25IQVIAYY650 pKa = 7.03TEE652 pKa = 4.21KK653 pKa = 10.31PDD655 pKa = 3.52GSFEE659 pKa = 4.14AEE661 pKa = 4.31PGFEE665 pKa = 4.26TPVPLSALEE674 pKa = 4.82ADD676 pKa = 4.61NPWLDD681 pKa = 3.52LVLWKK686 pKa = 10.21WEE688 pKa = 3.68GGRR691 pKa = 11.84YY692 pKa = 9.43VSMTPDD698 pKa = 3.25DD699 pKa = 3.91AAAVLLRR706 pKa = 11.84NRR708 pKa = 11.84GYY710 pKa = 9.53WVNARR715 pKa = 11.84RR716 pKa = 11.84GNICLVFPGSARR728 pKa = 11.84KK729 pKa = 9.3DD730 pKa = 3.55LPEE733 pKa = 6.28SEE735 pKa = 4.72DD736 pKa = 3.55TLATMMRR743 pKa = 11.84RR744 pKa = 11.84GGEE747 pKa = 3.54WMGRR751 pKa = 11.84HH752 pKa = 5.64LTPDD756 pKa = 3.16AAVADD761 pKa = 4.23SGEE764 pKa = 4.34TPPMPIGYY772 pKa = 8.82SGSGASGTGGGVDD785 pKa = 3.08SGGGDD790 pKa = 3.35VLLTRR795 pKa = 5.11
MM1 pKa = 7.49TSGEE5 pKa = 4.48YY6 pKa = 8.87FTVRR10 pKa = 11.84YY11 pKa = 9.28EE12 pKa = 4.07SGGVKK17 pKa = 10.16LVVSEE22 pKa = 4.71VISEE26 pKa = 3.86FSIGLTISPSGSGRR40 pKa = 11.84VLIEE44 pKa = 3.86EE45 pKa = 4.52EE46 pKa = 4.46GDD48 pKa = 3.8GEE50 pKa = 4.53TGTTGLTFTCPDD62 pKa = 3.76DD63 pKa = 5.52CVDD66 pKa = 5.77SDD68 pKa = 4.34TEE70 pKa = 4.43CNSGFCKK77 pKa = 10.36KK78 pKa = 10.17YY79 pKa = 9.06FAPGARR85 pKa = 11.84LKK87 pKa = 10.0LTATPGSGYY96 pKa = 10.75LFDD99 pKa = 3.57TWDD102 pKa = 3.98GEE104 pKa = 4.66VEE106 pKa = 4.32STSDD110 pKa = 3.41PKK112 pKa = 10.61IVYY115 pKa = 10.22VDD117 pKa = 3.68MAADD121 pKa = 3.4QSVTARR127 pKa = 11.84FEE129 pKa = 3.96ISQPLPDD136 pKa = 4.6RR137 pKa = 11.84DD138 pKa = 3.9GDD140 pKa = 4.15GTPDD144 pKa = 3.69TQDD147 pKa = 3.22GCPDD151 pKa = 3.98DD152 pKa = 4.16PRR154 pKa = 11.84KK155 pKa = 8.59TSPGACGCGVADD167 pKa = 4.16TDD169 pKa = 4.27SDD171 pKa = 3.91GDD173 pKa = 3.87EE174 pKa = 4.57TPDD177 pKa = 5.25CNDD180 pKa = 3.7EE181 pKa = 4.98CPADD185 pKa = 3.99PNKK188 pKa = 9.52TAPGQCGCGVADD200 pKa = 4.02TDD202 pKa = 4.27SDD204 pKa = 3.91GDD206 pKa = 3.87EE207 pKa = 4.57TPDD210 pKa = 5.29CNDD213 pKa = 3.9EE214 pKa = 5.15CPDD217 pKa = 4.71DD218 pKa = 4.48PDD220 pKa = 3.64KK221 pKa = 11.16TSPGACGCGVADD233 pKa = 4.24TDD235 pKa = 4.16SDD237 pKa = 3.8GDD239 pKa = 3.88ATPDD243 pKa = 3.93CNDD246 pKa = 3.99RR247 pKa = 11.84CPDD250 pKa = 4.29DD251 pKa = 4.24PDD253 pKa = 3.42KK254 pKa = 11.38TEE256 pKa = 5.01PGQCGCGLSDD266 pKa = 4.99ADD268 pKa = 3.92SDD270 pKa = 4.09GDD272 pKa = 4.02GFSDD276 pKa = 5.87CRR278 pKa = 11.84DD279 pKa = 3.41QCPDD283 pKa = 4.01DD284 pKa = 4.49PGKK287 pKa = 8.32TLPGQCGCGVADD299 pKa = 4.43TDD301 pKa = 3.74TDD303 pKa = 3.93GDD305 pKa = 4.01GTADD309 pKa = 4.9CNDD312 pKa = 4.02KK313 pKa = 11.53CPDD316 pKa = 4.16DD317 pKa = 4.26PNKK320 pKa = 9.94TFPGQCGCGVVDD332 pKa = 4.29TDD334 pKa = 4.36SDD336 pKa = 4.18GDD338 pKa = 3.93GTPDD342 pKa = 5.32CFDD345 pKa = 4.08KK346 pKa = 11.3CPNDD350 pKa = 3.66ADD352 pKa = 3.77KK353 pKa = 11.11TEE355 pKa = 4.89PGQCGCGVADD365 pKa = 4.99ADD367 pKa = 4.32SDD369 pKa = 4.29GDD371 pKa = 4.05GTPDD375 pKa = 5.26CSDD378 pKa = 3.76KK379 pKa = 11.35CPNDD383 pKa = 3.6PDD385 pKa = 3.59KK386 pKa = 11.31TEE388 pKa = 4.91PGQCGCGIADD398 pKa = 3.99TDD400 pKa = 3.85SDD402 pKa = 4.16GDD404 pKa = 4.02GVADD408 pKa = 4.17CEE410 pKa = 4.47SEE412 pKa = 4.29AVSGIPVPISPEE424 pKa = 3.69NGEE427 pKa = 4.32TVAEE431 pKa = 4.29DD432 pKa = 3.58SVTLVAQVDD441 pKa = 3.84LGSTVDD447 pKa = 4.58GPMEE451 pKa = 4.37IYY453 pKa = 8.94WWWRR457 pKa = 11.84PVDD460 pKa = 3.84KK461 pKa = 10.74KK462 pKa = 10.62CTVSSEE468 pKa = 3.98TTEE471 pKa = 4.08AYY473 pKa = 10.01RR474 pKa = 11.84GLAEE478 pKa = 4.58LRR480 pKa = 11.84ISGLRR485 pKa = 11.84DD486 pKa = 2.97GLQYY490 pKa = 11.13AWQVGVRR497 pKa = 11.84NPASGEE503 pKa = 3.8IFISDD508 pKa = 3.32EE509 pKa = 4.07VFFTVGASEE518 pKa = 4.42VEE520 pKa = 4.23VPFRR524 pKa = 11.84VDD526 pKa = 4.66AGTTLNDD533 pKa = 2.91YY534 pKa = 11.18RR535 pKa = 11.84MYY537 pKa = 10.98SFSLWPEE544 pKa = 4.04DD545 pKa = 4.45ADD547 pKa = 4.71AADD550 pKa = 4.27LLGDD554 pKa = 3.85AVGEE558 pKa = 4.03YY559 pKa = 8.24DD560 pKa = 3.72TKK562 pKa = 10.68FFKK565 pKa = 10.51IGTYY569 pKa = 10.36DD570 pKa = 3.67PEE572 pKa = 3.73QSRR575 pKa = 11.84YY576 pKa = 8.54VEE578 pKa = 4.15YY579 pKa = 10.72DD580 pKa = 2.7SGAITIVPGQAYY592 pKa = 7.88WFLARR597 pKa = 11.84NGLEE601 pKa = 3.68ASIRR605 pKa = 11.84GIKK608 pKa = 10.27VSVAEE613 pKa = 4.64NIHH616 pKa = 4.29VHH618 pKa = 5.8LRR620 pKa = 11.84YY621 pKa = 10.36DD622 pKa = 3.43EE623 pKa = 4.14TRR625 pKa = 11.84EE626 pKa = 4.15KK627 pKa = 9.22GWNMIAPPNDD637 pKa = 2.92GDD639 pKa = 4.03YY640 pKa = 10.67FRR642 pKa = 11.84KK643 pKa = 10.21DD644 pKa = 3.25IQVIAYY650 pKa = 7.03TEE652 pKa = 4.21KK653 pKa = 10.31PDD655 pKa = 3.52GSFEE659 pKa = 4.14AEE661 pKa = 4.31PGFEE665 pKa = 4.26TPVPLSALEE674 pKa = 4.82ADD676 pKa = 4.61NPWLDD681 pKa = 3.52LVLWKK686 pKa = 10.21WEE688 pKa = 3.68GGRR691 pKa = 11.84YY692 pKa = 9.43VSMTPDD698 pKa = 3.25DD699 pKa = 3.91AAAVLLRR706 pKa = 11.84NRR708 pKa = 11.84GYY710 pKa = 9.53WVNARR715 pKa = 11.84RR716 pKa = 11.84GNICLVFPGSARR728 pKa = 11.84KK729 pKa = 9.3DD730 pKa = 3.55LPEE733 pKa = 6.28SEE735 pKa = 4.72DD736 pKa = 3.55TLATMMRR743 pKa = 11.84RR744 pKa = 11.84GGEE747 pKa = 3.54WMGRR751 pKa = 11.84HH752 pKa = 5.64LTPDD756 pKa = 3.16AAVADD761 pKa = 4.23SGEE764 pKa = 4.34TPPMPIGYY772 pKa = 8.82SGSGASGTGGGVDD785 pKa = 3.08SGGGDD790 pKa = 3.35VLLTRR795 pKa = 5.11
Molecular weight: 84.04 kDa
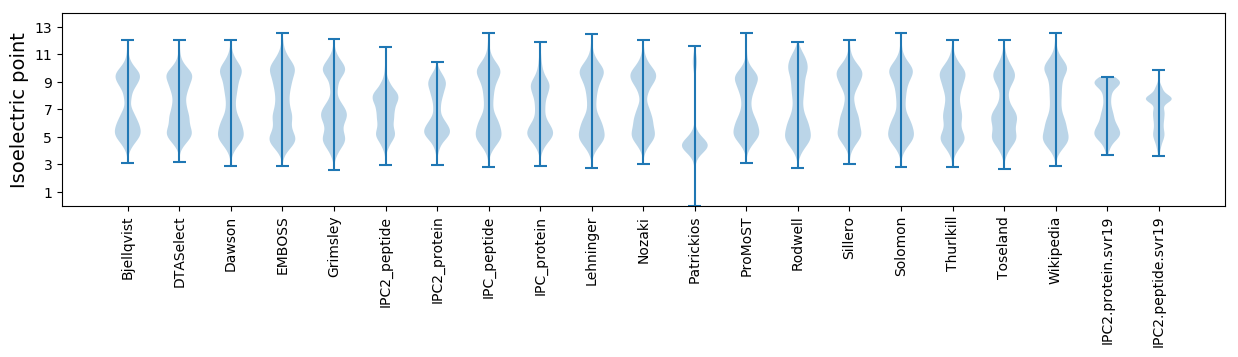
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401FSD9|A0A401FSD9_9DELT Ethanolamine ammonia-lyase OS=Desulfonema ishimotonii OX=45657 GN=DENIS_0833 PE=4 SV=1
MM1 pKa = 8.19AIRR4 pKa = 11.84KK5 pKa = 7.27IHH7 pKa = 6.69RR8 pKa = 11.84NITLWRR14 pKa = 11.84LTSDD18 pKa = 4.24FYY20 pKa = 11.68QFVRR24 pKa = 11.84SQRR27 pKa = 11.84FLTRR31 pKa = 11.84RR32 pKa = 11.84MGQPFSRR39 pKa = 11.84SRR41 pKa = 11.84KK42 pKa = 8.2FVEE45 pKa = 4.24MDD47 pKa = 2.99ITWRR51 pKa = 11.84CNLKK55 pKa = 10.64CPNCNRR61 pKa = 11.84SCTQVPSRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.63MSVGQVASFIRR82 pKa = 11.84EE83 pKa = 4.23SIAMGAQWEE92 pKa = 4.49RR93 pKa = 11.84IRR95 pKa = 11.84VLGGEE100 pKa = 4.3PTLHH104 pKa = 6.49RR105 pKa = 11.84GFFAILEE112 pKa = 4.28LLRR115 pKa = 11.84HH116 pKa = 5.51YY117 pKa = 10.34RR118 pKa = 11.84AAHH121 pKa = 5.33NPGLRR126 pKa = 11.84IVVCTNGHH134 pKa = 5.29GDD136 pKa = 3.29RR137 pKa = 11.84VNRR140 pKa = 11.84VLDD143 pKa = 4.99RR144 pKa = 11.84IPAGIAVKK152 pKa = 9.22NTFKK156 pKa = 10.49TAGPRR161 pKa = 11.84LFRR164 pKa = 11.84PFNVAPVDD172 pKa = 3.21THH174 pKa = 6.87RR175 pKa = 11.84YY176 pKa = 9.22RR177 pKa = 11.84FADD180 pKa = 3.4YY181 pKa = 11.15SGGCRR186 pKa = 11.84IISEE190 pKa = 4.4CGIGVTPLGCYY201 pKa = 9.14PCAIAGGIDD210 pKa = 3.52RR211 pKa = 11.84IFGFGLGRR219 pKa = 11.84KK220 pKa = 8.96KK221 pKa = 10.87LPLPDD226 pKa = 5.09DD227 pKa = 3.75EE228 pKa = 5.21MRR230 pKa = 11.84DD231 pKa = 3.5HH232 pKa = 7.15LKK234 pKa = 10.25IFCRR238 pKa = 11.84LCGHH242 pKa = 7.11FGFAWPTRR250 pKa = 11.84KK251 pKa = 9.71PEE253 pKa = 4.36ISKK256 pKa = 8.3TWEE259 pKa = 3.46NAYY262 pKa = 10.47RR263 pKa = 11.84NCRR266 pKa = 11.84AGNSSEE272 pKa = 4.0RR273 pKa = 3.74
MM1 pKa = 8.19AIRR4 pKa = 11.84KK5 pKa = 7.27IHH7 pKa = 6.69RR8 pKa = 11.84NITLWRR14 pKa = 11.84LTSDD18 pKa = 4.24FYY20 pKa = 11.68QFVRR24 pKa = 11.84SQRR27 pKa = 11.84FLTRR31 pKa = 11.84RR32 pKa = 11.84MGQPFSRR39 pKa = 11.84SRR41 pKa = 11.84KK42 pKa = 8.2FVEE45 pKa = 4.24MDD47 pKa = 2.99ITWRR51 pKa = 11.84CNLKK55 pKa = 10.64CPNCNRR61 pKa = 11.84SCTQVPSRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.63MSVGQVASFIRR82 pKa = 11.84EE83 pKa = 4.23SIAMGAQWEE92 pKa = 4.49RR93 pKa = 11.84IRR95 pKa = 11.84VLGGEE100 pKa = 4.3PTLHH104 pKa = 6.49RR105 pKa = 11.84GFFAILEE112 pKa = 4.28LLRR115 pKa = 11.84HH116 pKa = 5.51YY117 pKa = 10.34RR118 pKa = 11.84AAHH121 pKa = 5.33NPGLRR126 pKa = 11.84IVVCTNGHH134 pKa = 5.29GDD136 pKa = 3.29RR137 pKa = 11.84VNRR140 pKa = 11.84VLDD143 pKa = 4.99RR144 pKa = 11.84IPAGIAVKK152 pKa = 9.22NTFKK156 pKa = 10.49TAGPRR161 pKa = 11.84LFRR164 pKa = 11.84PFNVAPVDD172 pKa = 3.21THH174 pKa = 6.87RR175 pKa = 11.84YY176 pKa = 9.22RR177 pKa = 11.84FADD180 pKa = 3.4YY181 pKa = 11.15SGGCRR186 pKa = 11.84IISEE190 pKa = 4.4CGIGVTPLGCYY201 pKa = 9.14PCAIAGGIDD210 pKa = 3.52RR211 pKa = 11.84IFGFGLGRR219 pKa = 11.84KK220 pKa = 8.96KK221 pKa = 10.87LPLPDD226 pKa = 5.09DD227 pKa = 3.75EE228 pKa = 5.21MRR230 pKa = 11.84DD231 pKa = 3.5HH232 pKa = 7.15LKK234 pKa = 10.25IFCRR238 pKa = 11.84LCGHH242 pKa = 7.11FGFAWPTRR250 pKa = 11.84KK251 pKa = 9.71PEE253 pKa = 4.36ISKK256 pKa = 8.3TWEE259 pKa = 3.46NAYY262 pKa = 10.47RR263 pKa = 11.84NCRR266 pKa = 11.84AGNSSEE272 pKa = 4.0RR273 pKa = 3.74
Molecular weight: 31.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1708607 |
29 |
5505 |
340.9 |
37.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.708 ± 0.043 | 1.293 ± 0.015 |
5.892 ± 0.046 | 6.604 ± 0.031 |
4.347 ± 0.025 | 7.81 ± 0.036 |
1.993 ± 0.016 | 6.572 ± 0.032 |
5.113 ± 0.038 | 9.336 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.661 ± 0.018 | 3.473 ± 0.026 |
4.629 ± 0.026 | 2.988 ± 0.019 |
6.657 ± 0.041 | 5.733 ± 0.029 |
5.339 ± 0.04 | 6.645 ± 0.031 |
1.168 ± 0.014 | 3.038 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
