
Anaerobacillus arseniciselenatis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Anaerobacillus
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
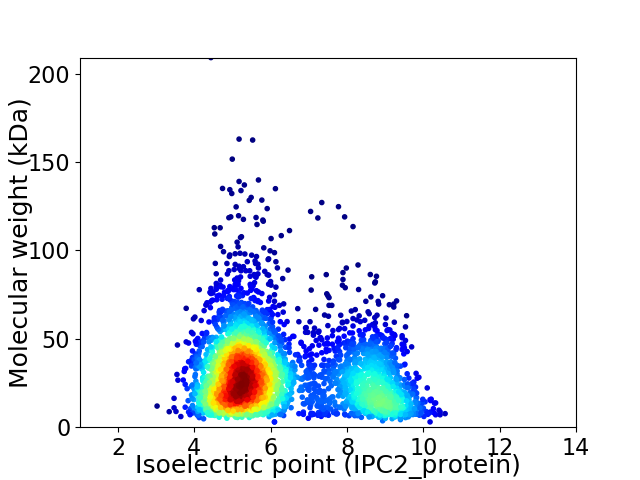
Virtual 2D-PAGE plot for 3587 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S2LQ82|A0A1S2LQ82_9BACI Uncharacterized protein OS=Anaerobacillus arseniciselenatis OX=85682 GN=BKP35_06265 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.18KK3 pKa = 9.77YY4 pKa = 10.72LKK6 pKa = 10.01RR7 pKa = 11.84LSIAAGLSLTLLAAGCGSDD26 pKa = 3.62ATTGDD31 pKa = 3.88DD32 pKa = 3.67SSVGGSLDD40 pKa = 3.57YY41 pKa = 10.95TITGIDD47 pKa = 3.18AGAGIMAATEE57 pKa = 4.03EE58 pKa = 4.56AIEE61 pKa = 4.42AYY63 pKa = 10.26GLEE66 pKa = 4.72DD67 pKa = 3.85YY68 pKa = 11.04NLQTSSSAAMTAALDD83 pKa = 3.53AAYY86 pKa = 9.32TNEE89 pKa = 3.9EE90 pKa = 4.59PIVVTGWTPHH100 pKa = 4.83WKK102 pKa = 9.0FAKK105 pKa = 10.28YY106 pKa = 9.77DD107 pKa = 3.76LKK109 pKa = 11.22YY110 pKa = 11.09LEE112 pKa = 5.21DD113 pKa = 3.82PQGIFGEE120 pKa = 4.53SEE122 pKa = 4.35VIHH125 pKa = 6.19TIARR129 pKa = 11.84AGLDD133 pKa = 3.4TDD135 pKa = 4.33HH136 pKa = 7.36PSAYY140 pKa = 9.41QVLEE144 pKa = 3.93NFYY147 pKa = 10.43WEE149 pKa = 4.09QDD151 pKa = 3.37DD152 pKa = 4.31MGAIMIDD159 pKa = 3.82VNEE162 pKa = 4.5GADD165 pKa = 3.81PVDD168 pKa = 4.76AAADD172 pKa = 3.52WVAANQDD179 pKa = 4.03KK180 pKa = 10.75VSEE183 pKa = 4.19WTDD186 pKa = 3.52GVSHH190 pKa = 7.12VDD192 pKa = 3.14GDD194 pKa = 4.18QITLAFVAWDD204 pKa = 3.52SEE206 pKa = 4.26IASTHH211 pKa = 5.8MIGKK215 pKa = 8.69VLEE218 pKa = 4.95DD219 pKa = 2.59IGYY222 pKa = 9.74DD223 pKa = 3.5VEE225 pKa = 4.82LVSLEE230 pKa = 3.99AAGMWTAVATGSADD244 pKa = 3.88AIVAAWLPGTHH255 pKa = 6.5AAYY258 pKa = 10.94YY259 pKa = 10.18EE260 pKa = 4.27DD261 pKa = 3.98YY262 pKa = 11.08KK263 pKa = 11.45EE264 pKa = 4.85DD265 pKa = 4.92FIDD268 pKa = 4.76LGVNLEE274 pKa = 4.07GAGIGLVVPAYY285 pKa = 9.9MDD287 pKa = 3.41IDD289 pKa = 4.58SIEE292 pKa = 4.26DD293 pKa = 3.5LNNN296 pKa = 3.12
MM1 pKa = 7.5KK2 pKa = 10.18KK3 pKa = 9.77YY4 pKa = 10.72LKK6 pKa = 10.01RR7 pKa = 11.84LSIAAGLSLTLLAAGCGSDD26 pKa = 3.62ATTGDD31 pKa = 3.88DD32 pKa = 3.67SSVGGSLDD40 pKa = 3.57YY41 pKa = 10.95TITGIDD47 pKa = 3.18AGAGIMAATEE57 pKa = 4.03EE58 pKa = 4.56AIEE61 pKa = 4.42AYY63 pKa = 10.26GLEE66 pKa = 4.72DD67 pKa = 3.85YY68 pKa = 11.04NLQTSSSAAMTAALDD83 pKa = 3.53AAYY86 pKa = 9.32TNEE89 pKa = 3.9EE90 pKa = 4.59PIVVTGWTPHH100 pKa = 4.83WKK102 pKa = 9.0FAKK105 pKa = 10.28YY106 pKa = 9.77DD107 pKa = 3.76LKK109 pKa = 11.22YY110 pKa = 11.09LEE112 pKa = 5.21DD113 pKa = 3.82PQGIFGEE120 pKa = 4.53SEE122 pKa = 4.35VIHH125 pKa = 6.19TIARR129 pKa = 11.84AGLDD133 pKa = 3.4TDD135 pKa = 4.33HH136 pKa = 7.36PSAYY140 pKa = 9.41QVLEE144 pKa = 3.93NFYY147 pKa = 10.43WEE149 pKa = 4.09QDD151 pKa = 3.37DD152 pKa = 4.31MGAIMIDD159 pKa = 3.82VNEE162 pKa = 4.5GADD165 pKa = 3.81PVDD168 pKa = 4.76AAADD172 pKa = 3.52WVAANQDD179 pKa = 4.03KK180 pKa = 10.75VSEE183 pKa = 4.19WTDD186 pKa = 3.52GVSHH190 pKa = 7.12VDD192 pKa = 3.14GDD194 pKa = 4.18QITLAFVAWDD204 pKa = 3.52SEE206 pKa = 4.26IASTHH211 pKa = 5.8MIGKK215 pKa = 8.69VLEE218 pKa = 4.95DD219 pKa = 2.59IGYY222 pKa = 9.74DD223 pKa = 3.5VEE225 pKa = 4.82LVSLEE230 pKa = 3.99AAGMWTAVATGSADD244 pKa = 3.88AIVAAWLPGTHH255 pKa = 6.5AAYY258 pKa = 10.94YY259 pKa = 10.18EE260 pKa = 4.27DD261 pKa = 3.98YY262 pKa = 11.08KK263 pKa = 11.45EE264 pKa = 4.85DD265 pKa = 4.92FIDD268 pKa = 4.76LGVNLEE274 pKa = 4.07GAGIGLVVPAYY285 pKa = 9.9MDD287 pKa = 3.41IDD289 pKa = 4.58SIEE292 pKa = 4.26DD293 pKa = 3.5LNNN296 pKa = 3.12
Molecular weight: 31.54 kDa
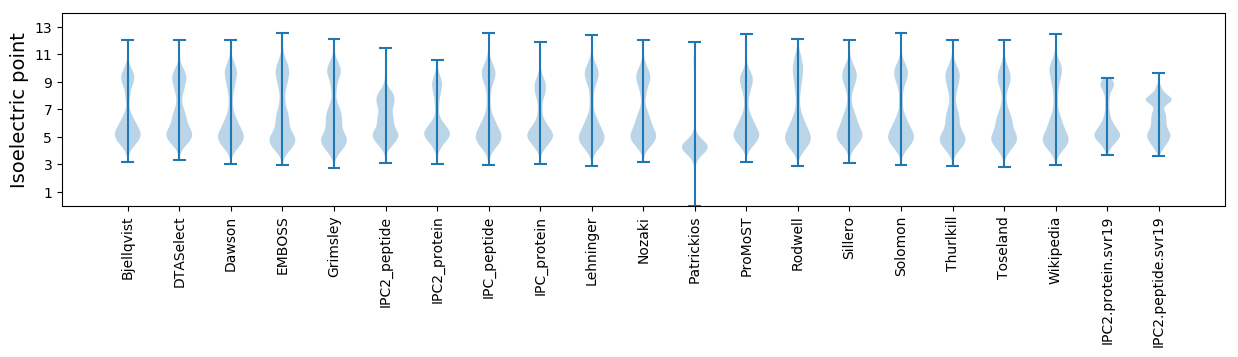
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S2LIU4|A0A1S2LIU4_9BACI Uncharacterized protein OS=Anaerobacillus arseniciselenatis OX=85682 GN=BKP35_10905 PE=4 SV=1
MM1 pKa = 6.37TTTVRR6 pKa = 11.84RR7 pKa = 11.84LVFFTALIAIWEE19 pKa = 4.64SIFRR23 pKa = 11.84LNDD26 pKa = 2.5IFTFRR31 pKa = 11.84DD32 pKa = 3.24TRR34 pKa = 11.84FFPSPSGAIEE44 pKa = 3.62QLYY47 pKa = 10.45IGFFQSGILSSALITSLQRR66 pKa = 11.84ISVGFALALIIGTALGILLGQSKK89 pKa = 10.09LADD92 pKa = 3.62EE93 pKa = 5.22TIGSLVIALQSIPSIVWLPIALIAFGQGSTAIIFIIVLGGTWAMTMNVRR142 pKa = 11.84MGIKK146 pKa = 9.74NVQPLLIRR154 pKa = 11.84AARR157 pKa = 11.84TMGYY161 pKa = 9.79KK162 pKa = 10.42GSEE165 pKa = 4.29LVWKK169 pKa = 10.59VMLPASIPAALTGARR184 pKa = 11.84LAWAFGWRR192 pKa = 11.84ALMAGEE198 pKa = 3.83LLGRR202 pKa = 11.84GGLGRR207 pKa = 11.84TLLDD211 pKa = 3.53ARR213 pKa = 11.84DD214 pKa = 4.25MFNMDD219 pKa = 3.95LVIAIMVIISIIGLIVEE236 pKa = 4.44YY237 pKa = 10.72LVFAPLEE244 pKa = 4.35KK245 pKa = 10.24KK246 pKa = 10.87VMTRR250 pKa = 11.84WGLSKK255 pKa = 10.97
MM1 pKa = 6.37TTTVRR6 pKa = 11.84RR7 pKa = 11.84LVFFTALIAIWEE19 pKa = 4.64SIFRR23 pKa = 11.84LNDD26 pKa = 2.5IFTFRR31 pKa = 11.84DD32 pKa = 3.24TRR34 pKa = 11.84FFPSPSGAIEE44 pKa = 3.62QLYY47 pKa = 10.45IGFFQSGILSSALITSLQRR66 pKa = 11.84ISVGFALALIIGTALGILLGQSKK89 pKa = 10.09LADD92 pKa = 3.62EE93 pKa = 5.22TIGSLVIALQSIPSIVWLPIALIAFGQGSTAIIFIIVLGGTWAMTMNVRR142 pKa = 11.84MGIKK146 pKa = 9.74NVQPLLIRR154 pKa = 11.84AARR157 pKa = 11.84TMGYY161 pKa = 9.79KK162 pKa = 10.42GSEE165 pKa = 4.29LVWKK169 pKa = 10.59VMLPASIPAALTGARR184 pKa = 11.84LAWAFGWRR192 pKa = 11.84ALMAGEE198 pKa = 3.83LLGRR202 pKa = 11.84GGLGRR207 pKa = 11.84TLLDD211 pKa = 3.53ARR213 pKa = 11.84DD214 pKa = 4.25MFNMDD219 pKa = 3.95LVIAIMVIISIIGLIVEE236 pKa = 4.44YY237 pKa = 10.72LVFAPLEE244 pKa = 4.35KK245 pKa = 10.24KK246 pKa = 10.87VMTRR250 pKa = 11.84WGLSKK255 pKa = 10.97
Molecular weight: 27.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1023233 |
26 |
1847 |
285.3 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.456 ± 0.05 | 0.789 ± 0.014 |
5.097 ± 0.033 | 7.877 ± 0.05 |
4.756 ± 0.04 | 6.589 ± 0.041 |
2.048 ± 0.02 | 8.131 ± 0.046 |
7.045 ± 0.049 | 9.732 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.659 ± 0.019 | 4.739 ± 0.033 |
3.46 ± 0.029 | 3.687 ± 0.029 |
3.94 ± 0.031 | 5.886 ± 0.031 |
5.398 ± 0.03 | 7.255 ± 0.041 |
0.971 ± 0.015 | 3.486 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
