
Kluyvera genomosp. 3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Kluyvera
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
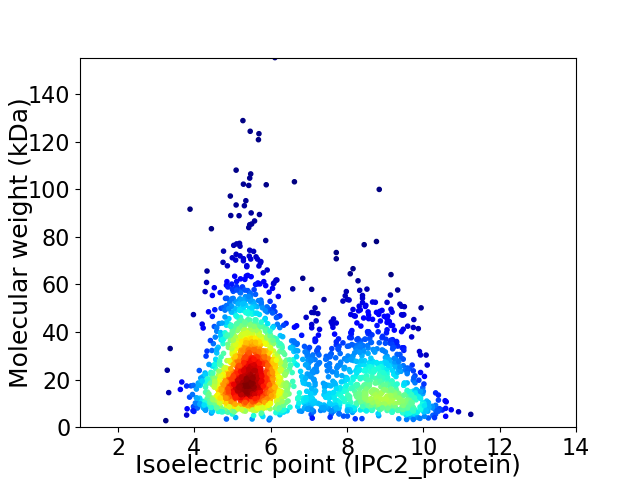
Virtual 2D-PAGE plot for 2234 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A248KHL4|A0A248KHL4_9ENTR Nitrate reductase molybdenum cofactor assembly chaperone OS=Kluyvera genomosp. 3 OX=2774055 GN=narJ PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84HH3 pKa = 4.86TARR6 pKa = 11.84LTLLSQCILLSLSAISGSALADD28 pKa = 3.25QTDD31 pKa = 4.06PCTGTSTTTCGFQDD45 pKa = 3.57KK46 pKa = 10.16TSTGPNGTSLIFVNNNGSAIMSDD69 pKa = 3.31AQNSNAIYY77 pKa = 10.04LWDD80 pKa = 3.53QTAGDD85 pKa = 4.12TQSLTVNGTDD95 pKa = 3.1MSGTYY100 pKa = 9.6IQGGYY105 pKa = 9.83IGTKK109 pKa = 10.42NITLNNATTDD119 pKa = 3.56MIEE122 pKa = 4.49AGNHH126 pKa = 6.46DD127 pKa = 4.67SGDD130 pKa = 3.75STNVNLAINNSTLNGEE146 pKa = 4.78DD147 pKa = 4.31DD148 pKa = 3.79STAYY152 pKa = 10.24GYY154 pKa = 11.03KK155 pKa = 9.33PAKK158 pKa = 9.52GNKK161 pKa = 9.65AYY163 pKa = 9.87MDD165 pKa = 4.22GAALFVDD172 pKa = 4.11SGSNAGTNNISIKK185 pKa = 10.62NGSSLMGSVYY195 pKa = 10.82AVTGGDD201 pKa = 3.62NNISMSDD208 pKa = 3.37SSIGGTNGSTGAIYY222 pKa = 10.88AMSNGGNNTITLEE235 pKa = 3.95NSTVVGSASEE245 pKa = 4.2PTDD248 pKa = 3.39KK249 pKa = 10.89TLLKK253 pKa = 10.61YY254 pKa = 10.84FEE256 pKa = 5.77DD257 pKa = 4.46NISGNSNASTIDD269 pKa = 3.26NLLNGSTIAMGVSGTQASSVALSNSKK295 pKa = 8.52VTGDD299 pKa = 2.97IAMVGTGSSSTASLNLSGNSNVTGDD324 pKa = 4.03ILLADD329 pKa = 4.31HH330 pKa = 6.73SAATVSMSDD339 pKa = 3.03STLTGNIDD347 pKa = 3.31ATNEE351 pKa = 3.8GNTAVALNNATVNGNITTGTGNDD374 pKa = 4.11SITLANNSHH383 pKa = 4.96VTGTVDD389 pKa = 3.47GGTGADD395 pKa = 3.95TLSLDD400 pKa = 3.98AGSSVDD406 pKa = 3.51GSIAQFEE413 pKa = 4.73TVNTAGNNSLTVDD426 pKa = 4.16TIEE429 pKa = 6.54DD430 pKa = 3.49NTTWNIQSGSTLFVTNTTGSNVQVNMSSDD459 pKa = 3.44SLVNLGTVGATANSNLVVSNTSMSTANQQNLAIATYY495 pKa = 7.21TTSASNPPNAVSVAFSNGAQQVEE518 pKa = 4.61SRR520 pKa = 11.84NGAYY524 pKa = 10.25NYY526 pKa = 10.67NNSLTQQAQPSTVSNGLLQANNDD549 pKa = 3.23TVYY552 pKa = 11.4NVMFSSSRR560 pKa = 11.84GDD562 pKa = 3.27LASDD566 pKa = 3.67VQGMIAGLDD575 pKa = 3.41AAKK578 pKa = 10.3QAGRR582 pKa = 11.84MITDD586 pKa = 3.95DD587 pKa = 3.62LANRR591 pKa = 11.84LTQVHH596 pKa = 6.14LQNLFGHH603 pKa = 6.44GVDD606 pKa = 4.9GAQVWGDD613 pKa = 3.37FLYY616 pKa = 11.17QNGDD620 pKa = 3.27YY621 pKa = 11.03SDD623 pKa = 4.45DD624 pKa = 3.52VDD626 pKa = 4.88YY627 pKa = 11.63KK628 pKa = 11.26DD629 pKa = 3.24ITQGVQGGVDD639 pKa = 3.21WTTHH643 pKa = 5.26LNNGDD648 pKa = 3.99SLTGGIALGWTRR660 pKa = 11.84SRR662 pKa = 11.84DD663 pKa = 3.17RR664 pKa = 11.84STNGGSNNFNDD675 pKa = 3.87SVYY678 pKa = 10.0GNYY681 pKa = 10.18YY682 pKa = 9.9SVYY685 pKa = 10.54GGWQQSLHH693 pKa = 6.54DD694 pKa = 4.68NLWGMFVDD702 pKa = 5.66GSFSYY707 pKa = 11.74GDD709 pKa = 3.37MRR711 pKa = 11.84YY712 pKa = 8.29STSANNVSNATTGMTQALDD731 pKa = 3.75GSSDD735 pKa = 3.4GNLYY739 pKa = 6.87TTQARR744 pKa = 11.84AGVNVVLPGEE754 pKa = 4.47TVIQPYY760 pKa = 8.31ATLGWDD766 pKa = 3.32KK767 pKa = 11.07AQEE770 pKa = 4.56DD771 pKa = 4.83GFSDD775 pKa = 3.55QAITFGDD782 pKa = 3.87SQVSEE787 pKa = 4.3WNTGIGMRR795 pKa = 11.84VTTKK799 pKa = 10.76LADD802 pKa = 3.73LNKK805 pKa = 10.33NVEE808 pKa = 4.32LYY810 pKa = 9.19PWLDD814 pKa = 2.81ARR816 pKa = 11.84YY817 pKa = 7.97QTEE820 pKa = 4.03FSDD823 pKa = 3.66NTDD826 pKa = 2.63IKK828 pKa = 11.07AADD831 pKa = 3.48YY832 pKa = 11.27HH833 pKa = 5.71NTNGHH838 pKa = 6.08NATMGIFGAGINTTIGKK855 pKa = 9.5DD856 pKa = 3.48FSLNTGVYY864 pKa = 10.06FGTGDD869 pKa = 3.35VDD871 pKa = 3.65NDD873 pKa = 3.34ASVQAGVSYY882 pKa = 10.85HH883 pKa = 5.72FF884 pKa = 4.73
MM1 pKa = 7.64RR2 pKa = 11.84HH3 pKa = 4.86TARR6 pKa = 11.84LTLLSQCILLSLSAISGSALADD28 pKa = 3.25QTDD31 pKa = 4.06PCTGTSTTTCGFQDD45 pKa = 3.57KK46 pKa = 10.16TSTGPNGTSLIFVNNNGSAIMSDD69 pKa = 3.31AQNSNAIYY77 pKa = 10.04LWDD80 pKa = 3.53QTAGDD85 pKa = 4.12TQSLTVNGTDD95 pKa = 3.1MSGTYY100 pKa = 9.6IQGGYY105 pKa = 9.83IGTKK109 pKa = 10.42NITLNNATTDD119 pKa = 3.56MIEE122 pKa = 4.49AGNHH126 pKa = 6.46DD127 pKa = 4.67SGDD130 pKa = 3.75STNVNLAINNSTLNGEE146 pKa = 4.78DD147 pKa = 4.31DD148 pKa = 3.79STAYY152 pKa = 10.24GYY154 pKa = 11.03KK155 pKa = 9.33PAKK158 pKa = 9.52GNKK161 pKa = 9.65AYY163 pKa = 9.87MDD165 pKa = 4.22GAALFVDD172 pKa = 4.11SGSNAGTNNISIKK185 pKa = 10.62NGSSLMGSVYY195 pKa = 10.82AVTGGDD201 pKa = 3.62NNISMSDD208 pKa = 3.37SSIGGTNGSTGAIYY222 pKa = 10.88AMSNGGNNTITLEE235 pKa = 3.95NSTVVGSASEE245 pKa = 4.2PTDD248 pKa = 3.39KK249 pKa = 10.89TLLKK253 pKa = 10.61YY254 pKa = 10.84FEE256 pKa = 5.77DD257 pKa = 4.46NISGNSNASTIDD269 pKa = 3.26NLLNGSTIAMGVSGTQASSVALSNSKK295 pKa = 8.52VTGDD299 pKa = 2.97IAMVGTGSSSTASLNLSGNSNVTGDD324 pKa = 4.03ILLADD329 pKa = 4.31HH330 pKa = 6.73SAATVSMSDD339 pKa = 3.03STLTGNIDD347 pKa = 3.31ATNEE351 pKa = 3.8GNTAVALNNATVNGNITTGTGNDD374 pKa = 4.11SITLANNSHH383 pKa = 4.96VTGTVDD389 pKa = 3.47GGTGADD395 pKa = 3.95TLSLDD400 pKa = 3.98AGSSVDD406 pKa = 3.51GSIAQFEE413 pKa = 4.73TVNTAGNNSLTVDD426 pKa = 4.16TIEE429 pKa = 6.54DD430 pKa = 3.49NTTWNIQSGSTLFVTNTTGSNVQVNMSSDD459 pKa = 3.44SLVNLGTVGATANSNLVVSNTSMSTANQQNLAIATYY495 pKa = 7.21TTSASNPPNAVSVAFSNGAQQVEE518 pKa = 4.61SRR520 pKa = 11.84NGAYY524 pKa = 10.25NYY526 pKa = 10.67NNSLTQQAQPSTVSNGLLQANNDD549 pKa = 3.23TVYY552 pKa = 11.4NVMFSSSRR560 pKa = 11.84GDD562 pKa = 3.27LASDD566 pKa = 3.67VQGMIAGLDD575 pKa = 3.41AAKK578 pKa = 10.3QAGRR582 pKa = 11.84MITDD586 pKa = 3.95DD587 pKa = 3.62LANRR591 pKa = 11.84LTQVHH596 pKa = 6.14LQNLFGHH603 pKa = 6.44GVDD606 pKa = 4.9GAQVWGDD613 pKa = 3.37FLYY616 pKa = 11.17QNGDD620 pKa = 3.27YY621 pKa = 11.03SDD623 pKa = 4.45DD624 pKa = 3.52VDD626 pKa = 4.88YY627 pKa = 11.63KK628 pKa = 11.26DD629 pKa = 3.24ITQGVQGGVDD639 pKa = 3.21WTTHH643 pKa = 5.26LNNGDD648 pKa = 3.99SLTGGIALGWTRR660 pKa = 11.84SRR662 pKa = 11.84DD663 pKa = 3.17RR664 pKa = 11.84STNGGSNNFNDD675 pKa = 3.87SVYY678 pKa = 10.0GNYY681 pKa = 10.18YY682 pKa = 9.9SVYY685 pKa = 10.54GGWQQSLHH693 pKa = 6.54DD694 pKa = 4.68NLWGMFVDD702 pKa = 5.66GSFSYY707 pKa = 11.74GDD709 pKa = 3.37MRR711 pKa = 11.84YY712 pKa = 8.29STSANNVSNATTGMTQALDD731 pKa = 3.75GSSDD735 pKa = 3.4GNLYY739 pKa = 6.87TTQARR744 pKa = 11.84AGVNVVLPGEE754 pKa = 4.47TVIQPYY760 pKa = 8.31ATLGWDD766 pKa = 3.32KK767 pKa = 11.07AQEE770 pKa = 4.56DD771 pKa = 4.83GFSDD775 pKa = 3.55QAITFGDD782 pKa = 3.87SQVSEE787 pKa = 4.3WNTGIGMRR795 pKa = 11.84VTTKK799 pKa = 10.76LADD802 pKa = 3.73LNKK805 pKa = 10.33NVEE808 pKa = 4.32LYY810 pKa = 9.19PWLDD814 pKa = 2.81ARR816 pKa = 11.84YY817 pKa = 7.97QTEE820 pKa = 4.03FSDD823 pKa = 3.66NTDD826 pKa = 2.63IKK828 pKa = 11.07AADD831 pKa = 3.48YY832 pKa = 11.27HH833 pKa = 5.71NTNGHH838 pKa = 6.08NATMGIFGAGINTTIGKK855 pKa = 9.5DD856 pKa = 3.48FSLNTGVYY864 pKa = 10.06FGTGDD869 pKa = 3.35VDD871 pKa = 3.65NDD873 pKa = 3.34ASVQAGVSYY882 pKa = 10.85HH883 pKa = 5.72FF884 pKa = 4.73
Molecular weight: 91.62 kDa
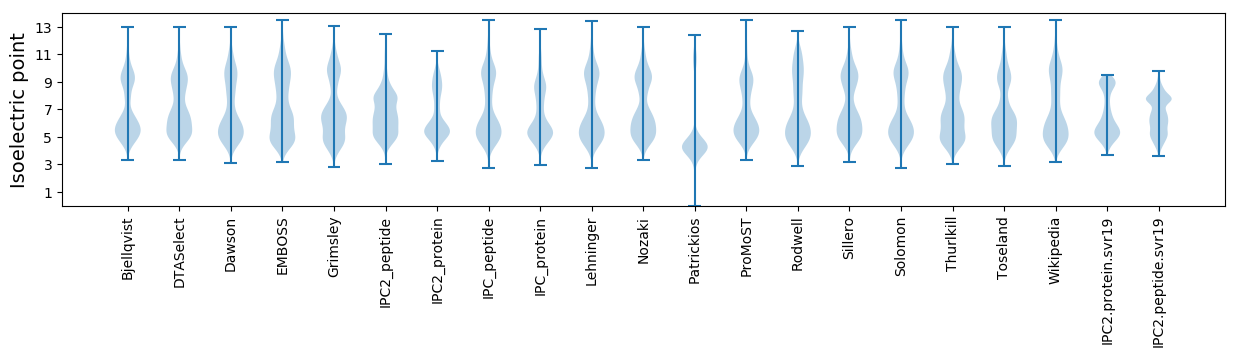
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A248KKU6|A0A248KKU6_9ENTR Peptide transporter OS=Kluyvera genomosp. 3 OX=2774055 GN=CEW81_19245 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84SRR41 pKa = 11.84LTVSKK46 pKa = 11.03
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84SRR41 pKa = 11.84LTVSKK46 pKa = 11.03
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
514278 |
26 |
1407 |
230.2 |
25.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.785 ± 0.067 | 1.142 ± 0.021 |
5.374 ± 0.043 | 5.734 ± 0.055 |
3.835 ± 0.043 | 7.255 ± 0.052 |
2.281 ± 0.028 | 5.8 ± 0.05 |
4.554 ± 0.049 | 10.293 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.824 ± 0.028 | 3.959 ± 0.042 |
4.134 ± 0.037 | 4.443 ± 0.042 |
5.588 ± 0.05 | 5.992 ± 0.046 |
5.542 ± 0.04 | 7.179 ± 0.048 |
1.401 ± 0.023 | 2.886 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
