
Dragonfly larvae associated circular virus-1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
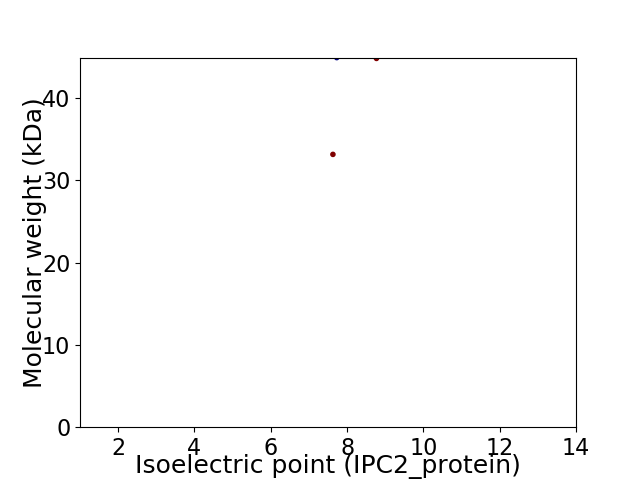
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
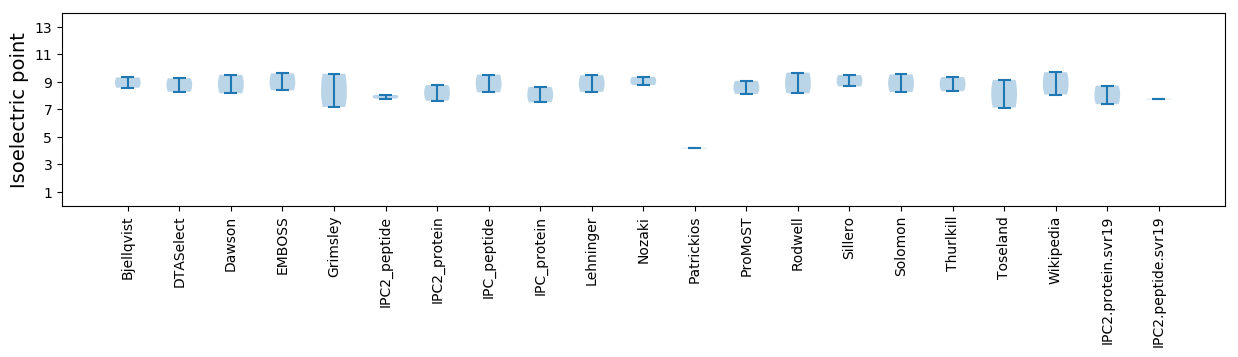
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U268|W5U268_9VIRU ATP-dependent helicase Rep OS=Dragonfly larvae associated circular virus-1 OX=1454021 PE=3 SV=1
MM1 pKa = 7.79PLSDD5 pKa = 4.51HH6 pKa = 7.2RR7 pKa = 11.84SPAPGSRR14 pKa = 11.84LSVRR18 pKa = 11.84RR19 pKa = 11.84QGIFWMLTIPHH30 pKa = 6.75HH31 pKa = 6.68CFTPYY36 pKa = 10.62LPPGCVWIRR45 pKa = 11.84GQHH48 pKa = 5.08EE49 pKa = 4.62LSLSGYY55 pKa = 6.95SHH57 pKa = 6.18WQILVAMSTKK67 pKa = 10.55SSLAQVKK74 pKa = 10.04SVFGEE79 pKa = 3.93QCHH82 pKa = 6.35AEE84 pKa = 4.15LSRR87 pKa = 11.84SEE89 pKa = 4.24SAQDD93 pKa = 3.78YY94 pKa = 8.97VWKK97 pKa = 10.35EE98 pKa = 3.95DD99 pKa = 3.43TRR101 pKa = 11.84VDD103 pKa = 3.55CSQFEE108 pKa = 4.47FGSKK112 pKa = 9.95PIRR115 pKa = 11.84RR116 pKa = 11.84NSRR119 pKa = 11.84VDD121 pKa = 3.23WEE123 pKa = 4.97SVWSAAKK130 pKa = 10.59SGDD133 pKa = 3.72LLQIPANVRR142 pKa = 11.84VVSYY146 pKa = 8.57RR147 pKa = 11.84TLRR150 pKa = 11.84AIKK153 pKa = 10.08ADD155 pKa = 3.72YY156 pKa = 9.16EE157 pKa = 4.81VPQSMEE163 pKa = 3.94RR164 pKa = 11.84TALCYY169 pKa = 9.29WGPTGTGKK177 pKa = 10.08SLRR180 pKa = 11.84SSRR183 pKa = 11.84EE184 pKa = 3.7AGIDD188 pKa = 3.46AYY190 pKa = 11.5YY191 pKa = 10.71KK192 pKa = 10.52DD193 pKa = 3.98PRR195 pKa = 11.84TKK197 pKa = 10.0FWCGYY202 pKa = 9.47RR203 pKa = 11.84GQQHH207 pKa = 6.08VIVDD211 pKa = 4.07EE212 pKa = 4.14FRR214 pKa = 11.84GAIDD218 pKa = 3.24ISHH221 pKa = 7.84LLRR224 pKa = 11.84WLDD227 pKa = 3.73RR228 pKa = 11.84YY229 pKa = 10.14PCYY232 pKa = 10.82LEE234 pKa = 4.38TKK236 pKa = 10.31GSSTSNCVTKK246 pKa = 10.42FWFTSNLHH254 pKa = 6.18PDD256 pKa = 3.67FWYY259 pKa = 10.14PDD261 pKa = 3.66LDD263 pKa = 3.77EE264 pKa = 4.57LTRR267 pKa = 11.84LALIRR272 pKa = 11.84RR273 pKa = 11.84ISVLEE278 pKa = 3.67ITEE281 pKa = 5.04LDD283 pKa = 3.57TSLLPP288 pKa = 4.48
MM1 pKa = 7.79PLSDD5 pKa = 4.51HH6 pKa = 7.2RR7 pKa = 11.84SPAPGSRR14 pKa = 11.84LSVRR18 pKa = 11.84RR19 pKa = 11.84QGIFWMLTIPHH30 pKa = 6.75HH31 pKa = 6.68CFTPYY36 pKa = 10.62LPPGCVWIRR45 pKa = 11.84GQHH48 pKa = 5.08EE49 pKa = 4.62LSLSGYY55 pKa = 6.95SHH57 pKa = 6.18WQILVAMSTKK67 pKa = 10.55SSLAQVKK74 pKa = 10.04SVFGEE79 pKa = 3.93QCHH82 pKa = 6.35AEE84 pKa = 4.15LSRR87 pKa = 11.84SEE89 pKa = 4.24SAQDD93 pKa = 3.78YY94 pKa = 8.97VWKK97 pKa = 10.35EE98 pKa = 3.95DD99 pKa = 3.43TRR101 pKa = 11.84VDD103 pKa = 3.55CSQFEE108 pKa = 4.47FGSKK112 pKa = 9.95PIRR115 pKa = 11.84RR116 pKa = 11.84NSRR119 pKa = 11.84VDD121 pKa = 3.23WEE123 pKa = 4.97SVWSAAKK130 pKa = 10.59SGDD133 pKa = 3.72LLQIPANVRR142 pKa = 11.84VVSYY146 pKa = 8.57RR147 pKa = 11.84TLRR150 pKa = 11.84AIKK153 pKa = 10.08ADD155 pKa = 3.72YY156 pKa = 9.16EE157 pKa = 4.81VPQSMEE163 pKa = 3.94RR164 pKa = 11.84TALCYY169 pKa = 9.29WGPTGTGKK177 pKa = 10.08SLRR180 pKa = 11.84SSRR183 pKa = 11.84EE184 pKa = 3.7AGIDD188 pKa = 3.46AYY190 pKa = 11.5YY191 pKa = 10.71KK192 pKa = 10.52DD193 pKa = 3.98PRR195 pKa = 11.84TKK197 pKa = 10.0FWCGYY202 pKa = 9.47RR203 pKa = 11.84GQQHH207 pKa = 6.08VIVDD211 pKa = 4.07EE212 pKa = 4.14FRR214 pKa = 11.84GAIDD218 pKa = 3.24ISHH221 pKa = 7.84LLRR224 pKa = 11.84WLDD227 pKa = 3.73RR228 pKa = 11.84YY229 pKa = 10.14PCYY232 pKa = 10.82LEE234 pKa = 4.38TKK236 pKa = 10.31GSSTSNCVTKK246 pKa = 10.42FWFTSNLHH254 pKa = 6.18PDD256 pKa = 3.67FWYY259 pKa = 10.14PDD261 pKa = 3.66LDD263 pKa = 3.77EE264 pKa = 4.57LTRR267 pKa = 11.84LALIRR272 pKa = 11.84RR273 pKa = 11.84ISVLEE278 pKa = 3.67ITEE281 pKa = 5.04LDD283 pKa = 3.57TSLLPP288 pKa = 4.48
Molecular weight: 33.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U268|W5U268_9VIRU ATP-dependent helicase Rep OS=Dragonfly larvae associated circular virus-1 OX=1454021 PE=3 SV=1
MM1 pKa = 7.38TRR3 pKa = 11.84TFNYY7 pKa = 8.73GHH9 pKa = 7.55PLRR12 pKa = 11.84VVGTIAANQFYY23 pKa = 10.73NKK25 pKa = 10.1YY26 pKa = 8.3IAPWKK31 pKa = 9.88DD32 pKa = 3.01RR33 pKa = 11.84KK34 pKa = 10.57HH35 pKa = 6.47PDD37 pKa = 2.65WLSYY41 pKa = 10.92KK42 pKa = 10.1PIDD45 pKa = 3.47YY46 pKa = 10.91SSIYY50 pKa = 10.39KK51 pKa = 9.31PDD53 pKa = 4.14HH54 pKa = 6.17GSKK57 pKa = 10.34PLPKK61 pKa = 9.97KK62 pKa = 10.59KK63 pKa = 9.92EE64 pKa = 4.01STMGAEE70 pKa = 5.17DD71 pKa = 3.52IPIGVKK77 pKa = 10.12KK78 pKa = 10.6NVVSVHH84 pKa = 5.91TGLKK88 pKa = 9.45RR89 pKa = 11.84LKK91 pKa = 10.82GMVAGHH97 pKa = 6.17ISYY100 pKa = 10.38RR101 pKa = 11.84DD102 pKa = 3.55SYY104 pKa = 10.72PALLVCDD111 pKa = 4.12EE112 pKa = 5.08GYY114 pKa = 11.18QNVTAPVYY122 pKa = 10.55IGTTSQFLTATSNGVASNRR141 pKa = 11.84DD142 pKa = 3.59SVSYY146 pKa = 10.97ASWFALNPEE155 pKa = 3.96QGIAAGSIISANTAPACDD173 pKa = 4.11KK174 pKa = 10.9IGLQSSTTYY183 pKa = 11.33LDD185 pKa = 3.9FKK187 pKa = 11.42NDD189 pKa = 3.52TTNICYY195 pKa = 10.22YY196 pKa = 10.17KK197 pKa = 10.0VHH199 pKa = 8.01IYY201 pKa = 7.98MAKK204 pKa = 8.86TDD206 pKa = 3.74TNQTVLSEE214 pKa = 4.5FNNSTGSDD222 pKa = 2.96NLYY225 pKa = 10.69NSNFAYY231 pKa = 8.55GTGVGTVLPATSGNEE246 pKa = 3.59QTNAYY251 pKa = 8.25IASSTSQIEE260 pKa = 4.45SCNMLPYY267 pKa = 10.32TKK269 pKa = 9.69ISSKK273 pKa = 10.59RR274 pKa = 11.84LVRR277 pKa = 11.84SVWKK281 pKa = 10.03KK282 pKa = 9.87IKK284 pKa = 9.58TISHH288 pKa = 5.74VFSGGDD294 pKa = 2.94AWKK297 pKa = 10.5LIVNIKK303 pKa = 10.05HH304 pKa = 5.35NQFGLRR310 pKa = 11.84EE311 pKa = 3.99RR312 pKa = 11.84LNTLSVVFPKK322 pKa = 10.96GCVCFLFEE330 pKa = 4.2MQGGTVYY337 pKa = 9.51NTANSAFGYY346 pKa = 8.2SACQMAVNITRR357 pKa = 11.84KK358 pKa = 9.08IHH360 pKa = 6.35LSPMAIPNKK369 pKa = 9.43RR370 pKa = 11.84FEE372 pKa = 4.1SRR374 pKa = 11.84YY375 pKa = 8.87VGAGRR380 pKa = 11.84EE381 pKa = 4.11FQGALLGTEE390 pKa = 3.7KK391 pKa = 10.81DD392 pKa = 3.59LYY394 pKa = 11.26HH395 pKa = 5.62NTEE398 pKa = 3.99VGAVPAKK405 pKa = 10.29IVV407 pKa = 3.05
MM1 pKa = 7.38TRR3 pKa = 11.84TFNYY7 pKa = 8.73GHH9 pKa = 7.55PLRR12 pKa = 11.84VVGTIAANQFYY23 pKa = 10.73NKK25 pKa = 10.1YY26 pKa = 8.3IAPWKK31 pKa = 9.88DD32 pKa = 3.01RR33 pKa = 11.84KK34 pKa = 10.57HH35 pKa = 6.47PDD37 pKa = 2.65WLSYY41 pKa = 10.92KK42 pKa = 10.1PIDD45 pKa = 3.47YY46 pKa = 10.91SSIYY50 pKa = 10.39KK51 pKa = 9.31PDD53 pKa = 4.14HH54 pKa = 6.17GSKK57 pKa = 10.34PLPKK61 pKa = 9.97KK62 pKa = 10.59KK63 pKa = 9.92EE64 pKa = 4.01STMGAEE70 pKa = 5.17DD71 pKa = 3.52IPIGVKK77 pKa = 10.12KK78 pKa = 10.6NVVSVHH84 pKa = 5.91TGLKK88 pKa = 9.45RR89 pKa = 11.84LKK91 pKa = 10.82GMVAGHH97 pKa = 6.17ISYY100 pKa = 10.38RR101 pKa = 11.84DD102 pKa = 3.55SYY104 pKa = 10.72PALLVCDD111 pKa = 4.12EE112 pKa = 5.08GYY114 pKa = 11.18QNVTAPVYY122 pKa = 10.55IGTTSQFLTATSNGVASNRR141 pKa = 11.84DD142 pKa = 3.59SVSYY146 pKa = 10.97ASWFALNPEE155 pKa = 3.96QGIAAGSIISANTAPACDD173 pKa = 4.11KK174 pKa = 10.9IGLQSSTTYY183 pKa = 11.33LDD185 pKa = 3.9FKK187 pKa = 11.42NDD189 pKa = 3.52TTNICYY195 pKa = 10.22YY196 pKa = 10.17KK197 pKa = 10.0VHH199 pKa = 8.01IYY201 pKa = 7.98MAKK204 pKa = 8.86TDD206 pKa = 3.74TNQTVLSEE214 pKa = 4.5FNNSTGSDD222 pKa = 2.96NLYY225 pKa = 10.69NSNFAYY231 pKa = 8.55GTGVGTVLPATSGNEE246 pKa = 3.59QTNAYY251 pKa = 8.25IASSTSQIEE260 pKa = 4.45SCNMLPYY267 pKa = 10.32TKK269 pKa = 9.69ISSKK273 pKa = 10.59RR274 pKa = 11.84LVRR277 pKa = 11.84SVWKK281 pKa = 10.03KK282 pKa = 9.87IKK284 pKa = 9.58TISHH288 pKa = 5.74VFSGGDD294 pKa = 2.94AWKK297 pKa = 10.5LIVNIKK303 pKa = 10.05HH304 pKa = 5.35NQFGLRR310 pKa = 11.84EE311 pKa = 3.99RR312 pKa = 11.84LNTLSVVFPKK322 pKa = 10.96GCVCFLFEE330 pKa = 4.2MQGGTVYY337 pKa = 9.51NTANSAFGYY346 pKa = 8.2SACQMAVNITRR357 pKa = 11.84KK358 pKa = 9.08IHH360 pKa = 6.35LSPMAIPNKK369 pKa = 9.43RR370 pKa = 11.84FEE372 pKa = 4.1SRR374 pKa = 11.84YY375 pKa = 8.87VGAGRR380 pKa = 11.84EE381 pKa = 4.11FQGALLGTEE390 pKa = 3.7KK391 pKa = 10.81DD392 pKa = 3.59LYY394 pKa = 11.26HH395 pKa = 5.62NTEE398 pKa = 3.99VGAVPAKK405 pKa = 10.29IVV407 pKa = 3.05
Molecular weight: 44.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
695 |
288 |
407 |
347.5 |
38.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.619 ± 0.868 | 2.158 ± 0.381 |
4.46 ± 0.674 | 4.029 ± 0.726 |
3.597 ± 0.077 | 6.763 ± 0.743 |
2.734 ± 0.241 | 5.612 ± 0.462 |
5.755 ± 1.191 | 7.482 ± 1.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.727 ± 0.208 | 4.604 ± 1.978 |
4.892 ± 0.408 | 3.309 ± 0.314 |
5.468 ± 1.763 | 9.928 ± 0.728 |
6.906 ± 0.831 | 6.619 ± 0.44 |
2.302 ± 0.933 | 5.036 ± 0.535 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
