
Coraliomargarita sinensis
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Opitutae; Puniceicoccales; Puniceicoccaceae; Coraliomargarita
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
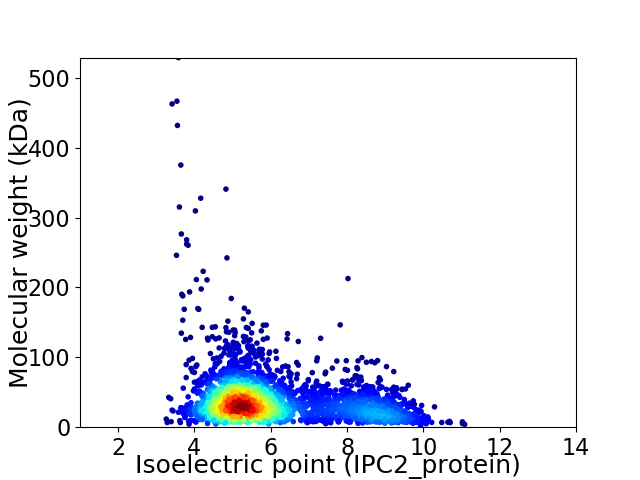
Virtual 2D-PAGE plot for 2981 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
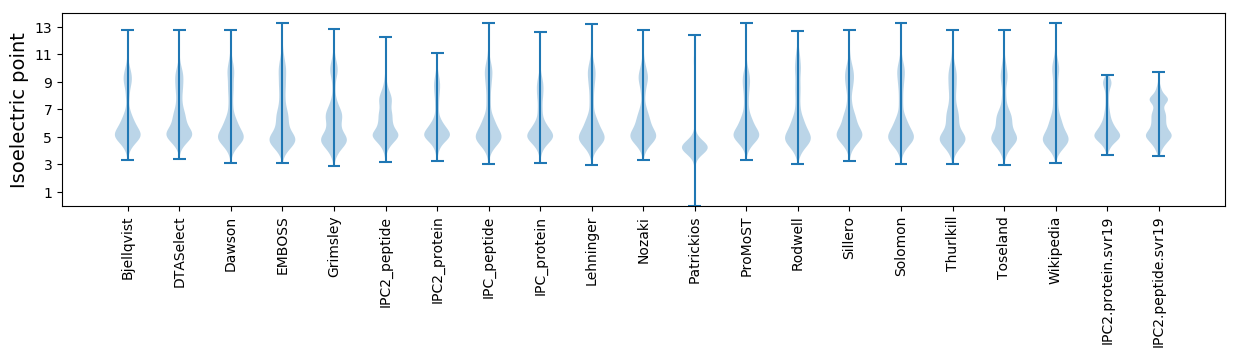
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317ZDZ7|A0A317ZDZ7_9BACT Putative pre-16S rRNA nuclease OS=Coraliomargarita sinensis OX=2174842 GN=DDZ13_14855 PE=3 SV=1
MM1 pKa = 7.2QKK3 pKa = 10.13NAPICLALAVCYY15 pKa = 7.37LTWLAPLRR23 pKa = 11.84AADD26 pKa = 3.42ITFVGHH32 pKa = 5.82QDD34 pKa = 3.35AVEE37 pKa = 4.37ADD39 pKa = 3.51ADD41 pKa = 4.27TPTSGWRR48 pKa = 11.84NATPSKK54 pKa = 9.89TLDD57 pKa = 3.16IDD59 pKa = 3.51VDD61 pKa = 3.99NVLGTDD67 pKa = 4.73GYY69 pKa = 11.13QVAAKK74 pKa = 9.73SANPSYY80 pKa = 10.92GWIIRR85 pKa = 11.84RR86 pKa = 11.84SPGYY90 pKa = 10.03NHH92 pKa = 7.14ASSWGLWDD100 pKa = 5.44DD101 pKa = 4.16PDD103 pKa = 3.92NASGSDD109 pKa = 3.48NLSAGVFYY117 pKa = 10.78DD118 pKa = 4.04PNANGQVDD126 pKa = 4.02VFEE129 pKa = 4.76LVIEE133 pKa = 4.29GDD135 pKa = 3.74EE136 pKa = 4.37LVGKK140 pKa = 6.6TLRR143 pKa = 11.84VGILYY148 pKa = 10.38GVMVNGTSTFTVTQTVGGNQMVTTPSLTHH177 pKa = 7.25DD178 pKa = 3.57ATSLNAAFFNITGAVEE194 pKa = 4.1GDD196 pKa = 3.49TFRR199 pKa = 11.84ISTEE203 pKa = 3.47ILTGSGVEE211 pKa = 3.93QVGGIFLDD219 pKa = 3.91SAPTNPGTQLPEE231 pKa = 3.28VDD233 pKa = 3.24GTVRR237 pKa = 11.84GTYY240 pKa = 8.81VAPLEE245 pKa = 4.26VAGIFSDD252 pKa = 3.18NGVIQRR258 pKa = 11.84GMVVPVWGWDD268 pKa = 3.5DD269 pKa = 4.02PGTVVTVDD277 pKa = 3.8FAGQSVNGTADD288 pKa = 3.23ANGRR292 pKa = 11.84WEE294 pKa = 4.63AEE296 pKa = 4.16LLSLTANTTPQTMTISAGADD316 pKa = 2.85TVTISNLLVGDD327 pKa = 3.88VWLCSGQSNMEE338 pKa = 3.65MDD340 pKa = 4.43FNGSQYY346 pKa = 10.97AADD349 pKa = 3.94MPAFIDD355 pKa = 3.81PDD357 pKa = 3.5NFPTIRR363 pKa = 11.84HH364 pKa = 5.89IDD366 pKa = 3.24ISNAQAEE373 pKa = 4.65FRR375 pKa = 11.84QKK377 pKa = 10.76RR378 pKa = 11.84LPNGNAGWTVCDD390 pKa = 3.39STTVQDD396 pKa = 3.46YY397 pKa = 8.21TAAGFFFATTISQEE411 pKa = 3.96TGVPIGLINSTFGNSRR427 pKa = 11.84IGEE430 pKa = 4.32FVTPDD435 pKa = 3.45SIGLVTDD442 pKa = 4.09QIPASSFRR450 pKa = 11.84GVFTPDD456 pKa = 2.85PCKK459 pKa = 10.7KK460 pKa = 9.44FFAMVAPITGYY471 pKa = 11.74AMTGALWYY479 pKa = 10.41QGEE482 pKa = 4.24ANANDD487 pKa = 3.81GEE489 pKa = 4.55DD490 pKa = 4.16YY491 pKa = 10.36FWKK494 pKa = 10.47LAALFQGWRR503 pKa = 11.84DD504 pKa = 3.1EE505 pKa = 4.21WQQGDD510 pKa = 3.92FPFYY514 pKa = 10.21YY515 pKa = 10.58AQLPSFDD522 pKa = 5.1GSVSWPIIRR531 pKa = 11.84EE532 pKa = 4.08SQRR535 pKa = 11.84RR536 pKa = 11.84ALTLTNTGMATLVDD550 pKa = 3.86VGGVGLPWPEE560 pKa = 3.97NLHH563 pKa = 6.45PPNKK567 pKa = 9.44YY568 pKa = 10.09DD569 pKa = 3.13VGRR572 pKa = 11.84RR573 pKa = 11.84LAQWALLNEE582 pKa = 4.39YY583 pKa = 9.15GQSSVVPSGPLFSSAVFDD601 pKa = 4.04GANVTISFDD610 pKa = 3.77YY611 pKa = 10.97VGSGLVAAAKK621 pKa = 9.15PSPQSTDD628 pKa = 2.91APSPVADD635 pKa = 3.31VLGFEE640 pKa = 4.79LAGSDD645 pKa = 4.24EE646 pKa = 4.33VWHH649 pKa = 6.6AANASIAGSTVVLSSASVPNPEE671 pKa = 3.97AARR674 pKa = 11.84YY675 pKa = 9.18LYY677 pKa = 9.26ATNTDD682 pKa = 3.44GGTLYY687 pKa = 11.0NLEE690 pKa = 5.11DD691 pKa = 3.8LPAAPFLTTAADD703 pKa = 3.9NPNPPSTPTGLNATSGDD720 pKa = 3.48GRR722 pKa = 11.84VDD724 pKa = 4.15LAWDD728 pKa = 3.71AATGADD734 pKa = 3.58SYY736 pKa = 11.26NVKK739 pKa = 10.43RR740 pKa = 11.84STTTGGPYY748 pKa = 10.58ASIGSPSGTSYY759 pKa = 12.01ADD761 pKa = 3.15TTAANGTTYY770 pKa = 10.38YY771 pKa = 10.83YY772 pKa = 11.13VVAAVNTIGSSADD785 pKa = 3.36STEE788 pKa = 4.21VSATPVEE795 pKa = 4.52ATNPPDD801 pKa = 3.85APTGLSATPGDD812 pKa = 3.78NRR814 pKa = 11.84VDD816 pKa = 4.2LSWSASSGADD826 pKa = 2.95SYY828 pKa = 11.44NVKK831 pKa = 10.36RR832 pKa = 11.84STTMGGPYY840 pKa = 9.81SGIGTTPGISYY851 pKa = 10.13TDD853 pKa = 3.84LSATNGTTYY862 pKa = 10.69YY863 pKa = 10.56YY864 pKa = 10.83VVSASNTYY872 pKa = 10.6GNSADD877 pKa = 3.75SAEE880 pKa = 4.4ASATPQEE887 pKa = 4.32PAAGAVITYY896 pKa = 9.72VGHH899 pKa = 6.42QDD901 pKa = 3.45NVDD904 pKa = 3.5AKK906 pKa = 10.12TDD908 pKa = 3.72DD909 pKa = 3.88PTSGWRR915 pKa = 11.84NTTPAKK921 pKa = 9.78THH923 pKa = 7.22DD924 pKa = 3.6IDD926 pKa = 4.58GDD928 pKa = 4.16NILGTDD934 pKa = 4.47GYY936 pKa = 11.88AMFRR940 pKa = 11.84NNSAFSVVSLPSYY953 pKa = 10.18ISAVTHH959 pKa = 6.41VSTHH963 pKa = 5.65SNANASFGVMDD974 pKa = 4.63NPSDD978 pKa = 3.75PSGSDD983 pKa = 3.61DD984 pKa = 3.25INYY987 pKa = 9.25GLWYY991 pKa = 9.02LTTTGGTTDD1000 pKa = 3.18VFTFTITGTDD1010 pKa = 3.44LDD1012 pKa = 4.33GKK1014 pKa = 6.76TLRR1017 pKa = 11.84VGLAYY1022 pKa = 10.3DD1023 pKa = 4.88GYY1025 pKa = 10.86NGSGNQQLTITQTAGGAAADD1045 pKa = 3.83NSGTVSWGNDD1055 pKa = 2.85GLDD1058 pKa = 3.35FVFFDD1063 pKa = 3.3ITEE1066 pKa = 4.23VNDD1069 pKa = 3.53GDD1071 pKa = 4.26VFTVNASAGAGLPHH1085 pKa = 6.46VSGVTFDD1092 pKa = 4.32VITDD1096 pKa = 3.62IATPPGTPTGLAATPGDD1113 pKa = 3.9GQVDD1117 pKa = 4.21LTWNVSSGAASYY1129 pKa = 10.21NVKK1132 pKa = 10.43RR1133 pKa = 11.84GTAEE1137 pKa = 4.01GGPYY1141 pKa = 8.85STVASPTTNSFSDD1154 pKa = 3.6STVSNGTTYY1163 pKa = 10.73YY1164 pKa = 10.58YY1165 pKa = 10.9VVSAVNAEE1173 pKa = 4.19GEE1175 pKa = 4.5SPDD1178 pKa = 3.8SGEE1181 pKa = 5.04ASATPQALPPDD1192 pKa = 4.27APTGLVATPGDD1203 pKa = 3.99GQVDD1207 pKa = 4.5LSWDD1211 pKa = 3.46ASSGADD1217 pKa = 3.08SYY1219 pKa = 11.35NVKK1222 pKa = 10.28RR1223 pKa = 11.84GTSMGGPYY1231 pKa = 9.0STVASPNNNSLTDD1244 pKa = 3.54ATVSNGTTYY1253 pKa = 10.73YY1254 pKa = 10.58YY1255 pKa = 10.86VVSAVNTNGEE1265 pKa = 4.34SGNSGEE1271 pKa = 5.07VSATPQAAPTGATISYY1287 pKa = 9.94VGNQTNVEE1295 pKa = 4.34TEE1297 pKa = 3.78ASDD1300 pKa = 5.22AGIGWRR1306 pKa = 11.84NTTPDD1311 pKa = 3.75KK1312 pKa = 11.11PLDD1315 pKa = 3.58IDD1317 pKa = 3.95GDD1319 pKa = 4.29NILGTDD1325 pKa = 4.41GYY1327 pKa = 11.14RR1328 pKa = 11.84IFNDD1332 pKa = 3.55GSTVSLPSYY1341 pKa = 9.85IDD1343 pKa = 4.33NITKK1347 pKa = 10.22IAPNAAKK1354 pKa = 10.44SGGWGVIDD1362 pKa = 5.7DD1363 pKa = 4.82PSDD1366 pKa = 3.48PSGTDD1371 pKa = 3.69DD1372 pKa = 4.42IIHH1375 pKa = 6.12GFWYY1379 pKa = 10.08DD1380 pKa = 3.07SGNAQGSVDD1389 pKa = 3.26ILSFQITGTALDD1401 pKa = 4.19GQTLRR1406 pKa = 11.84IGIIYY1411 pKa = 7.63DD1412 pKa = 3.72THH1414 pKa = 6.66WGTGDD1419 pKa = 3.0IAFTLTQNGTSGSVTATSGLVTAGSDD1445 pKa = 3.75GIDD1448 pKa = 3.12VVFFDD1453 pKa = 4.64LTNVSSGDD1461 pKa = 3.76VFTLNADD1468 pKa = 3.65ADD1470 pKa = 4.0DD1471 pKa = 5.09HH1472 pKa = 6.38NFSHH1476 pKa = 7.23AGGITFDD1483 pKa = 3.52TSVISGGDD1491 pKa = 3.43TLADD1495 pKa = 3.2WLASPEE1501 pKa = 4.26FGLDD1505 pKa = 3.56PADD1508 pKa = 3.94QDD1510 pKa = 5.15FGDD1513 pKa = 4.93DD1514 pKa = 3.78PDD1516 pKa = 6.15GDD1518 pKa = 3.63GHH1520 pKa = 7.71ANGIEE1525 pKa = 4.41NYY1527 pKa = 10.05FGTHH1531 pKa = 6.45PGEE1534 pKa = 4.34FSQGLVSGTVSGDD1547 pKa = 2.96SMTFTHH1553 pKa = 7.16PLSDD1557 pKa = 3.96SPAADD1562 pKa = 2.99ISAVYY1567 pKa = 9.68RR1568 pKa = 11.84WSKK1571 pKa = 11.06DD1572 pKa = 2.92LDD1574 pKa = 3.88SFHH1577 pKa = 8.1VDD1579 pKa = 2.95GDD1581 pKa = 4.17SFDD1584 pKa = 3.73NTTVTFSQGTPADD1597 pKa = 3.48GMVEE1601 pKa = 3.82VTAAITGDD1609 pKa = 3.87PLDD1612 pKa = 4.14KK1613 pKa = 10.91LFVDD1617 pKa = 4.9IEE1619 pKa = 4.45VTQPP1623 pKa = 2.93
MM1 pKa = 7.2QKK3 pKa = 10.13NAPICLALAVCYY15 pKa = 7.37LTWLAPLRR23 pKa = 11.84AADD26 pKa = 3.42ITFVGHH32 pKa = 5.82QDD34 pKa = 3.35AVEE37 pKa = 4.37ADD39 pKa = 3.51ADD41 pKa = 4.27TPTSGWRR48 pKa = 11.84NATPSKK54 pKa = 9.89TLDD57 pKa = 3.16IDD59 pKa = 3.51VDD61 pKa = 3.99NVLGTDD67 pKa = 4.73GYY69 pKa = 11.13QVAAKK74 pKa = 9.73SANPSYY80 pKa = 10.92GWIIRR85 pKa = 11.84RR86 pKa = 11.84SPGYY90 pKa = 10.03NHH92 pKa = 7.14ASSWGLWDD100 pKa = 5.44DD101 pKa = 4.16PDD103 pKa = 3.92NASGSDD109 pKa = 3.48NLSAGVFYY117 pKa = 10.78DD118 pKa = 4.04PNANGQVDD126 pKa = 4.02VFEE129 pKa = 4.76LVIEE133 pKa = 4.29GDD135 pKa = 3.74EE136 pKa = 4.37LVGKK140 pKa = 6.6TLRR143 pKa = 11.84VGILYY148 pKa = 10.38GVMVNGTSTFTVTQTVGGNQMVTTPSLTHH177 pKa = 7.25DD178 pKa = 3.57ATSLNAAFFNITGAVEE194 pKa = 4.1GDD196 pKa = 3.49TFRR199 pKa = 11.84ISTEE203 pKa = 3.47ILTGSGVEE211 pKa = 3.93QVGGIFLDD219 pKa = 3.91SAPTNPGTQLPEE231 pKa = 3.28VDD233 pKa = 3.24GTVRR237 pKa = 11.84GTYY240 pKa = 8.81VAPLEE245 pKa = 4.26VAGIFSDD252 pKa = 3.18NGVIQRR258 pKa = 11.84GMVVPVWGWDD268 pKa = 3.5DD269 pKa = 4.02PGTVVTVDD277 pKa = 3.8FAGQSVNGTADD288 pKa = 3.23ANGRR292 pKa = 11.84WEE294 pKa = 4.63AEE296 pKa = 4.16LLSLTANTTPQTMTISAGADD316 pKa = 2.85TVTISNLLVGDD327 pKa = 3.88VWLCSGQSNMEE338 pKa = 3.65MDD340 pKa = 4.43FNGSQYY346 pKa = 10.97AADD349 pKa = 3.94MPAFIDD355 pKa = 3.81PDD357 pKa = 3.5NFPTIRR363 pKa = 11.84HH364 pKa = 5.89IDD366 pKa = 3.24ISNAQAEE373 pKa = 4.65FRR375 pKa = 11.84QKK377 pKa = 10.76RR378 pKa = 11.84LPNGNAGWTVCDD390 pKa = 3.39STTVQDD396 pKa = 3.46YY397 pKa = 8.21TAAGFFFATTISQEE411 pKa = 3.96TGVPIGLINSTFGNSRR427 pKa = 11.84IGEE430 pKa = 4.32FVTPDD435 pKa = 3.45SIGLVTDD442 pKa = 4.09QIPASSFRR450 pKa = 11.84GVFTPDD456 pKa = 2.85PCKK459 pKa = 10.7KK460 pKa = 9.44FFAMVAPITGYY471 pKa = 11.74AMTGALWYY479 pKa = 10.41QGEE482 pKa = 4.24ANANDD487 pKa = 3.81GEE489 pKa = 4.55DD490 pKa = 4.16YY491 pKa = 10.36FWKK494 pKa = 10.47LAALFQGWRR503 pKa = 11.84DD504 pKa = 3.1EE505 pKa = 4.21WQQGDD510 pKa = 3.92FPFYY514 pKa = 10.21YY515 pKa = 10.58AQLPSFDD522 pKa = 5.1GSVSWPIIRR531 pKa = 11.84EE532 pKa = 4.08SQRR535 pKa = 11.84RR536 pKa = 11.84ALTLTNTGMATLVDD550 pKa = 3.86VGGVGLPWPEE560 pKa = 3.97NLHH563 pKa = 6.45PPNKK567 pKa = 9.44YY568 pKa = 10.09DD569 pKa = 3.13VGRR572 pKa = 11.84RR573 pKa = 11.84LAQWALLNEE582 pKa = 4.39YY583 pKa = 9.15GQSSVVPSGPLFSSAVFDD601 pKa = 4.04GANVTISFDD610 pKa = 3.77YY611 pKa = 10.97VGSGLVAAAKK621 pKa = 9.15PSPQSTDD628 pKa = 2.91APSPVADD635 pKa = 3.31VLGFEE640 pKa = 4.79LAGSDD645 pKa = 4.24EE646 pKa = 4.33VWHH649 pKa = 6.6AANASIAGSTVVLSSASVPNPEE671 pKa = 3.97AARR674 pKa = 11.84YY675 pKa = 9.18LYY677 pKa = 9.26ATNTDD682 pKa = 3.44GGTLYY687 pKa = 11.0NLEE690 pKa = 5.11DD691 pKa = 3.8LPAAPFLTTAADD703 pKa = 3.9NPNPPSTPTGLNATSGDD720 pKa = 3.48GRR722 pKa = 11.84VDD724 pKa = 4.15LAWDD728 pKa = 3.71AATGADD734 pKa = 3.58SYY736 pKa = 11.26NVKK739 pKa = 10.43RR740 pKa = 11.84STTTGGPYY748 pKa = 10.58ASIGSPSGTSYY759 pKa = 12.01ADD761 pKa = 3.15TTAANGTTYY770 pKa = 10.38YY771 pKa = 10.83YY772 pKa = 11.13VVAAVNTIGSSADD785 pKa = 3.36STEE788 pKa = 4.21VSATPVEE795 pKa = 4.52ATNPPDD801 pKa = 3.85APTGLSATPGDD812 pKa = 3.78NRR814 pKa = 11.84VDD816 pKa = 4.2LSWSASSGADD826 pKa = 2.95SYY828 pKa = 11.44NVKK831 pKa = 10.36RR832 pKa = 11.84STTMGGPYY840 pKa = 9.81SGIGTTPGISYY851 pKa = 10.13TDD853 pKa = 3.84LSATNGTTYY862 pKa = 10.69YY863 pKa = 10.56YY864 pKa = 10.83VVSASNTYY872 pKa = 10.6GNSADD877 pKa = 3.75SAEE880 pKa = 4.4ASATPQEE887 pKa = 4.32PAAGAVITYY896 pKa = 9.72VGHH899 pKa = 6.42QDD901 pKa = 3.45NVDD904 pKa = 3.5AKK906 pKa = 10.12TDD908 pKa = 3.72DD909 pKa = 3.88PTSGWRR915 pKa = 11.84NTTPAKK921 pKa = 9.78THH923 pKa = 7.22DD924 pKa = 3.6IDD926 pKa = 4.58GDD928 pKa = 4.16NILGTDD934 pKa = 4.47GYY936 pKa = 11.88AMFRR940 pKa = 11.84NNSAFSVVSLPSYY953 pKa = 10.18ISAVTHH959 pKa = 6.41VSTHH963 pKa = 5.65SNANASFGVMDD974 pKa = 4.63NPSDD978 pKa = 3.75PSGSDD983 pKa = 3.61DD984 pKa = 3.25INYY987 pKa = 9.25GLWYY991 pKa = 9.02LTTTGGTTDD1000 pKa = 3.18VFTFTITGTDD1010 pKa = 3.44LDD1012 pKa = 4.33GKK1014 pKa = 6.76TLRR1017 pKa = 11.84VGLAYY1022 pKa = 10.3DD1023 pKa = 4.88GYY1025 pKa = 10.86NGSGNQQLTITQTAGGAAADD1045 pKa = 3.83NSGTVSWGNDD1055 pKa = 2.85GLDD1058 pKa = 3.35FVFFDD1063 pKa = 3.3ITEE1066 pKa = 4.23VNDD1069 pKa = 3.53GDD1071 pKa = 4.26VFTVNASAGAGLPHH1085 pKa = 6.46VSGVTFDD1092 pKa = 4.32VITDD1096 pKa = 3.62IATPPGTPTGLAATPGDD1113 pKa = 3.9GQVDD1117 pKa = 4.21LTWNVSSGAASYY1129 pKa = 10.21NVKK1132 pKa = 10.43RR1133 pKa = 11.84GTAEE1137 pKa = 4.01GGPYY1141 pKa = 8.85STVASPTTNSFSDD1154 pKa = 3.6STVSNGTTYY1163 pKa = 10.73YY1164 pKa = 10.58YY1165 pKa = 10.9VVSAVNAEE1173 pKa = 4.19GEE1175 pKa = 4.5SPDD1178 pKa = 3.8SGEE1181 pKa = 5.04ASATPQALPPDD1192 pKa = 4.27APTGLVATPGDD1203 pKa = 3.99GQVDD1207 pKa = 4.5LSWDD1211 pKa = 3.46ASSGADD1217 pKa = 3.08SYY1219 pKa = 11.35NVKK1222 pKa = 10.28RR1223 pKa = 11.84GTSMGGPYY1231 pKa = 9.0STVASPNNNSLTDD1244 pKa = 3.54ATVSNGTTYY1253 pKa = 10.73YY1254 pKa = 10.58YY1255 pKa = 10.86VVSAVNTNGEE1265 pKa = 4.34SGNSGEE1271 pKa = 5.07VSATPQAAPTGATISYY1287 pKa = 9.94VGNQTNVEE1295 pKa = 4.34TEE1297 pKa = 3.78ASDD1300 pKa = 5.22AGIGWRR1306 pKa = 11.84NTTPDD1311 pKa = 3.75KK1312 pKa = 11.11PLDD1315 pKa = 3.58IDD1317 pKa = 3.95GDD1319 pKa = 4.29NILGTDD1325 pKa = 4.41GYY1327 pKa = 11.14RR1328 pKa = 11.84IFNDD1332 pKa = 3.55GSTVSLPSYY1341 pKa = 9.85IDD1343 pKa = 4.33NITKK1347 pKa = 10.22IAPNAAKK1354 pKa = 10.44SGGWGVIDD1362 pKa = 5.7DD1363 pKa = 4.82PSDD1366 pKa = 3.48PSGTDD1371 pKa = 3.69DD1372 pKa = 4.42IIHH1375 pKa = 6.12GFWYY1379 pKa = 10.08DD1380 pKa = 3.07SGNAQGSVDD1389 pKa = 3.26ILSFQITGTALDD1401 pKa = 4.19GQTLRR1406 pKa = 11.84IGIIYY1411 pKa = 7.63DD1412 pKa = 3.72THH1414 pKa = 6.66WGTGDD1419 pKa = 3.0IAFTLTQNGTSGSVTATSGLVTAGSDD1445 pKa = 3.75GIDD1448 pKa = 3.12VVFFDD1453 pKa = 4.64LTNVSSGDD1461 pKa = 3.76VFTLNADD1468 pKa = 3.65ADD1470 pKa = 4.0DD1471 pKa = 5.09HH1472 pKa = 6.38NFSHH1476 pKa = 7.23AGGITFDD1483 pKa = 3.52TSVISGGDD1491 pKa = 3.43TLADD1495 pKa = 3.2WLASPEE1501 pKa = 4.26FGLDD1505 pKa = 3.56PADD1508 pKa = 3.94QDD1510 pKa = 5.15FGDD1513 pKa = 4.93DD1514 pKa = 3.78PDD1516 pKa = 6.15GDD1518 pKa = 3.63GHH1520 pKa = 7.71ANGIEE1525 pKa = 4.41NYY1527 pKa = 10.05FGTHH1531 pKa = 6.45PGEE1534 pKa = 4.34FSQGLVSGTVSGDD1547 pKa = 2.96SMTFTHH1553 pKa = 7.16PLSDD1557 pKa = 3.96SPAADD1562 pKa = 2.99ISAVYY1567 pKa = 9.68RR1568 pKa = 11.84WSKK1571 pKa = 11.06DD1572 pKa = 2.92LDD1574 pKa = 3.88SFHH1577 pKa = 8.1VDD1579 pKa = 2.95GDD1581 pKa = 4.17SFDD1584 pKa = 3.73NTTVTFSQGTPADD1597 pKa = 3.48GMVEE1601 pKa = 3.82VTAAITGDD1609 pKa = 3.87PLDD1612 pKa = 4.14KK1613 pKa = 10.91LFVDD1617 pKa = 4.9IEE1619 pKa = 4.45VTQPP1623 pKa = 2.93
Molecular weight: 168.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317ZD97|A0A317ZD97_9BACT Cytochrome oxidase assembly protein OS=Coraliomargarita sinensis OX=2174842 GN=DDZ13_13100 PE=4 SV=1
MM1 pKa = 7.59GNLKK5 pKa = 10.05KK6 pKa = 10.3KK7 pKa = 10.32RR8 pKa = 11.84RR9 pKa = 11.84LKK11 pKa = 9.51MNKK14 pKa = 8.33HH15 pKa = 5.49KK16 pKa = 10.35RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.17RR20 pKa = 11.84LKK22 pKa = 9.94SNRR25 pKa = 11.84HH26 pKa = 5.57KK27 pKa = 10.88KK28 pKa = 8.44RR29 pKa = 11.84TWW31 pKa = 2.8
MM1 pKa = 7.59GNLKK5 pKa = 10.05KK6 pKa = 10.3KK7 pKa = 10.32RR8 pKa = 11.84RR9 pKa = 11.84LKK11 pKa = 9.51MNKK14 pKa = 8.33HH15 pKa = 5.49KK16 pKa = 10.35RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.17RR20 pKa = 11.84LKK22 pKa = 9.94SNRR25 pKa = 11.84HH26 pKa = 5.57KK27 pKa = 10.88KK28 pKa = 8.44RR29 pKa = 11.84TWW31 pKa = 2.8
Molecular weight: 4.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1056829 |
15 |
5145 |
354.5 |
39.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.111 ± 0.049 | 1.019 ± 0.018 |
6.035 ± 0.055 | 7.15 ± 0.056 |
4.201 ± 0.03 | 7.778 ± 0.054 |
2.106 ± 0.028 | 5.691 ± 0.04 |
4.803 ± 0.066 | 9.752 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.18 ± 0.025 | 3.593 ± 0.036 |
4.712 ± 0.03 | 3.405 ± 0.028 |
5.716 ± 0.057 | 6.443 ± 0.051 |
5.301 ± 0.075 | 6.584 ± 0.04 |
1.405 ± 0.021 | 3.015 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
