
Wenxinia marina DSM 24838
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Wenxinia; Wenxinia marina
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
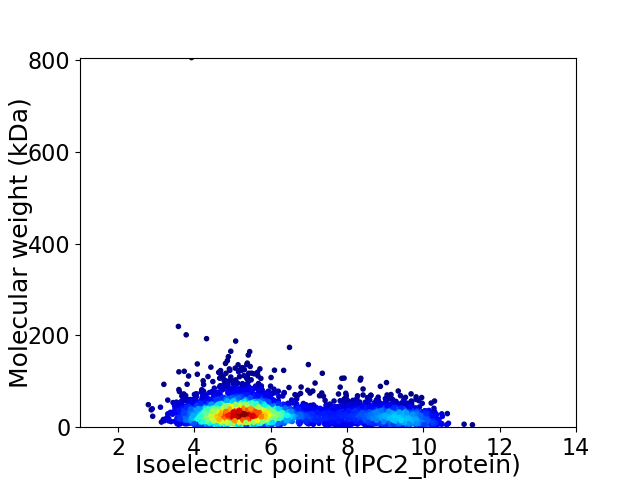
Virtual 2D-PAGE plot for 4030 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
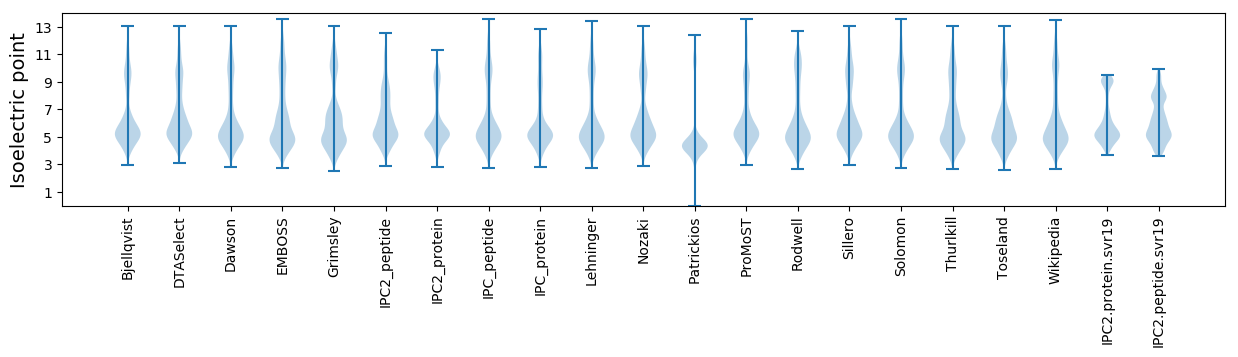
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D0Q8B7|A0A0D0Q8B7_9RHOB Mg chelatase-related protein OS=Wenxinia marina DSM 24838 OX=1123501 GN=Wenmar_01022 PE=3 SV=1
MM1 pKa = 7.2TADD4 pKa = 3.67NNTDD8 pKa = 3.35GEE10 pKa = 4.32IHH12 pKa = 6.51MKK14 pKa = 10.46RR15 pKa = 11.84LLLSSAALLLPAGTALADD33 pKa = 3.88FSMTILHH40 pKa = 6.41TNDD43 pKa = 2.48FHH45 pKa = 8.99DD46 pKa = 3.87RR47 pKa = 11.84FEE49 pKa = 5.48PISAFDD55 pKa = 3.7STCDD59 pKa = 3.71AEE61 pKa = 4.37TNAAGEE67 pKa = 4.58CFGGTARR74 pKa = 11.84LVTAIEE80 pKa = 3.75QARR83 pKa = 11.84AEE85 pKa = 4.18NEE87 pKa = 3.7NTILVDD93 pKa = 5.05GGDD96 pKa = 3.59QFQGTLFYY104 pKa = 10.03TYY106 pKa = 10.75YY107 pKa = 9.55KK108 pKa = 11.14GSMTAEE114 pKa = 4.31FMNQLGYY121 pKa = 10.99DD122 pKa = 3.29AMTVGNHH129 pKa = 5.62EE130 pKa = 4.45FDD132 pKa = 5.23DD133 pKa = 5.09GPEE136 pKa = 3.95VLRR139 pKa = 11.84EE140 pKa = 3.92FVDD143 pKa = 3.92TVEE146 pKa = 5.11FPILMSNADD155 pKa = 3.52VSGEE159 pKa = 4.03PLLADD164 pKa = 5.56AIMKK168 pKa = 7.98STTIEE173 pKa = 4.04VGGEE177 pKa = 4.12TIGLIGLTPQDD188 pKa = 3.73TDD190 pKa = 3.46EE191 pKa = 4.75LASPGPNVIFTDD203 pKa = 3.99PSDD206 pKa = 3.63AVQAEE211 pKa = 4.12VDD213 pKa = 3.57ALTEE217 pKa = 4.16AGVNKK222 pKa = 9.98IVVLSHH228 pKa = 6.42SGYY231 pKa = 11.36GLDD234 pKa = 3.37QTVAEE239 pKa = 4.49NTTGVDD245 pKa = 3.88VIVGGHH251 pKa = 5.65TNTLLGDD258 pKa = 3.52MEE260 pKa = 4.8GAQGAYY266 pKa = 7.34PTMVGDD272 pKa = 3.75TAIVQAYY279 pKa = 10.11AYY281 pKa = 9.78GKK283 pKa = 9.11YY284 pKa = 10.4LGMLDD289 pKa = 3.66VTFDD293 pKa = 3.72DD294 pKa = 4.99DD295 pKa = 4.71GNIIEE300 pKa = 5.6ASGQPLLIDD309 pKa = 3.91ASVEE313 pKa = 4.1EE314 pKa = 4.26NQGVLDD320 pKa = 4.6QIAEE324 pKa = 4.2YY325 pKa = 10.23AAPLDD330 pKa = 4.23EE331 pKa = 4.61IRR333 pKa = 11.84NEE335 pKa = 4.23VVAEE339 pKa = 3.89TSEE342 pKa = 4.67AIEE345 pKa = 4.34GNRR348 pKa = 11.84DD349 pKa = 2.33ICRR352 pKa = 11.84AMEE355 pKa = 4.17CSMGNLVADD364 pKa = 4.1AMLDD368 pKa = 3.4RR369 pKa = 11.84VADD372 pKa = 3.5QGIDD376 pKa = 3.43VAIANSGGLRR386 pKa = 11.84ASIDD390 pKa = 3.41AGEE393 pKa = 4.19VTMGEE398 pKa = 4.52VLTVLPFQNTLSTFQITGAQLMEE421 pKa = 4.32ALEE424 pKa = 4.5NGVSQVEE431 pKa = 4.34EE432 pKa = 4.13GAGRR436 pKa = 11.84FPQIAGMTFTADD448 pKa = 3.74LSQEE452 pKa = 3.9PGSRR456 pKa = 11.84ITEE459 pKa = 4.02VTVGGEE465 pKa = 4.02PLDD468 pKa = 4.17PEE470 pKa = 4.19ATYY473 pKa = 10.78GAVSNNYY480 pKa = 8.3VRR482 pKa = 11.84NGGDD486 pKa = 3.63GYY488 pKa = 11.6SMFVNAEE495 pKa = 3.72NAYY498 pKa = 10.81DD499 pKa = 4.41FGPDD503 pKa = 3.4LADD506 pKa = 3.24VTAEE510 pKa = 3.85YY511 pKa = 10.49LAANGPYY518 pKa = 9.49TPYY521 pKa = 9.58TDD523 pKa = 3.45GRR525 pKa = 11.84ITIEE529 pKa = 3.86NAPAAEE535 pKa = 4.02
MM1 pKa = 7.2TADD4 pKa = 3.67NNTDD8 pKa = 3.35GEE10 pKa = 4.32IHH12 pKa = 6.51MKK14 pKa = 10.46RR15 pKa = 11.84LLLSSAALLLPAGTALADD33 pKa = 3.88FSMTILHH40 pKa = 6.41TNDD43 pKa = 2.48FHH45 pKa = 8.99DD46 pKa = 3.87RR47 pKa = 11.84FEE49 pKa = 5.48PISAFDD55 pKa = 3.7STCDD59 pKa = 3.71AEE61 pKa = 4.37TNAAGEE67 pKa = 4.58CFGGTARR74 pKa = 11.84LVTAIEE80 pKa = 3.75QARR83 pKa = 11.84AEE85 pKa = 4.18NEE87 pKa = 3.7NTILVDD93 pKa = 5.05GGDD96 pKa = 3.59QFQGTLFYY104 pKa = 10.03TYY106 pKa = 10.75YY107 pKa = 9.55KK108 pKa = 11.14GSMTAEE114 pKa = 4.31FMNQLGYY121 pKa = 10.99DD122 pKa = 3.29AMTVGNHH129 pKa = 5.62EE130 pKa = 4.45FDD132 pKa = 5.23DD133 pKa = 5.09GPEE136 pKa = 3.95VLRR139 pKa = 11.84EE140 pKa = 3.92FVDD143 pKa = 3.92TVEE146 pKa = 5.11FPILMSNADD155 pKa = 3.52VSGEE159 pKa = 4.03PLLADD164 pKa = 5.56AIMKK168 pKa = 7.98STTIEE173 pKa = 4.04VGGEE177 pKa = 4.12TIGLIGLTPQDD188 pKa = 3.73TDD190 pKa = 3.46EE191 pKa = 4.75LASPGPNVIFTDD203 pKa = 3.99PSDD206 pKa = 3.63AVQAEE211 pKa = 4.12VDD213 pKa = 3.57ALTEE217 pKa = 4.16AGVNKK222 pKa = 9.98IVVLSHH228 pKa = 6.42SGYY231 pKa = 11.36GLDD234 pKa = 3.37QTVAEE239 pKa = 4.49NTTGVDD245 pKa = 3.88VIVGGHH251 pKa = 5.65TNTLLGDD258 pKa = 3.52MEE260 pKa = 4.8GAQGAYY266 pKa = 7.34PTMVGDD272 pKa = 3.75TAIVQAYY279 pKa = 10.11AYY281 pKa = 9.78GKK283 pKa = 9.11YY284 pKa = 10.4LGMLDD289 pKa = 3.66VTFDD293 pKa = 3.72DD294 pKa = 4.99DD295 pKa = 4.71GNIIEE300 pKa = 5.6ASGQPLLIDD309 pKa = 3.91ASVEE313 pKa = 4.1EE314 pKa = 4.26NQGVLDD320 pKa = 4.6QIAEE324 pKa = 4.2YY325 pKa = 10.23AAPLDD330 pKa = 4.23EE331 pKa = 4.61IRR333 pKa = 11.84NEE335 pKa = 4.23VVAEE339 pKa = 3.89TSEE342 pKa = 4.67AIEE345 pKa = 4.34GNRR348 pKa = 11.84DD349 pKa = 2.33ICRR352 pKa = 11.84AMEE355 pKa = 4.17CSMGNLVADD364 pKa = 4.1AMLDD368 pKa = 3.4RR369 pKa = 11.84VADD372 pKa = 3.5QGIDD376 pKa = 3.43VAIANSGGLRR386 pKa = 11.84ASIDD390 pKa = 3.41AGEE393 pKa = 4.19VTMGEE398 pKa = 4.52VLTVLPFQNTLSTFQITGAQLMEE421 pKa = 4.32ALEE424 pKa = 4.5NGVSQVEE431 pKa = 4.34EE432 pKa = 4.13GAGRR436 pKa = 11.84FPQIAGMTFTADD448 pKa = 3.74LSQEE452 pKa = 3.9PGSRR456 pKa = 11.84ITEE459 pKa = 4.02VTVGGEE465 pKa = 4.02PLDD468 pKa = 4.17PEE470 pKa = 4.19ATYY473 pKa = 10.78GAVSNNYY480 pKa = 8.3VRR482 pKa = 11.84NGGDD486 pKa = 3.63GYY488 pKa = 11.6SMFVNAEE495 pKa = 3.72NAYY498 pKa = 10.81DD499 pKa = 4.41FGPDD503 pKa = 3.4LADD506 pKa = 3.24VTAEE510 pKa = 3.85YY511 pKa = 10.49LAANGPYY518 pKa = 9.49TPYY521 pKa = 9.58TDD523 pKa = 3.45GRR525 pKa = 11.84ITIEE529 pKa = 3.86NAPAAEE535 pKa = 4.02
Molecular weight: 56.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D0Q484|A0A0D0Q484_9RHOB GTPase Der OS=Wenxinia marina DSM 24838 OX=1123501 GN=der PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84VRR13 pKa = 11.84KK14 pKa = 9.2ARR16 pKa = 11.84HH17 pKa = 4.71GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.43GGRR29 pKa = 11.84KK30 pKa = 8.99VLNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84VRR13 pKa = 11.84KK14 pKa = 9.2ARR16 pKa = 11.84HH17 pKa = 4.71GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.43GGRR29 pKa = 11.84KK30 pKa = 8.99VLNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1236898 |
24 |
7873 |
306.9 |
33.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.692 ± 0.06 | 0.808 ± 0.012 |
6.179 ± 0.044 | 6.101 ± 0.037 |
3.422 ± 0.027 | 9.382 ± 0.044 |
1.918 ± 0.02 | 4.508 ± 0.029 |
2.064 ± 0.033 | 10.178 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.494 ± 0.02 | 2.047 ± 0.022 |
5.604 ± 0.038 | 2.65 ± 0.02 |
7.781 ± 0.049 | 4.575 ± 0.032 |
5.413 ± 0.032 | 7.591 ± 0.036 |
1.534 ± 0.019 | 2.056 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
