
Salicibibacter kimchii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Salicibibacter
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
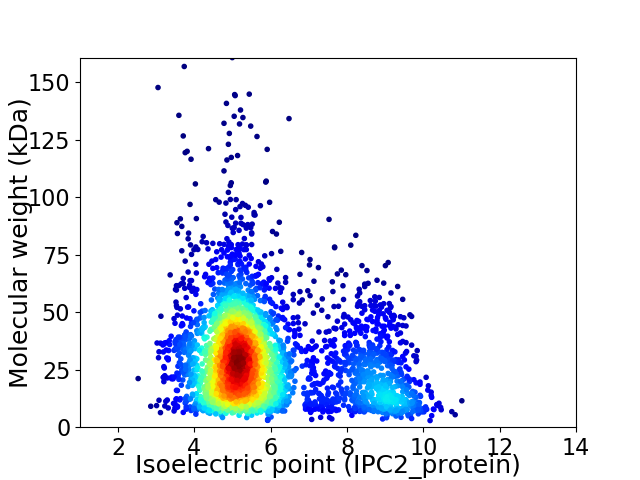
Virtual 2D-PAGE plot for 3509 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345C0U3|A0A345C0U3_9BACI Copper chaperone OS=Salicibibacter kimchii OX=2099786 GN=DT065_12965 PE=4 SV=1
MM1 pKa = 7.17KK2 pKa = 9.99KK3 pKa = 10.46VYY5 pKa = 10.19IGMSTIFFVLAGCSGEE21 pKa = 4.2GLDD24 pKa = 4.5EE25 pKa = 4.86GSTGSTEE32 pKa = 4.08SQQEE36 pKa = 4.25GKK38 pKa = 10.15GNEE41 pKa = 3.94MMTEE45 pKa = 3.91DD46 pKa = 3.5GKK48 pKa = 11.21EE49 pKa = 3.96VFNLDD54 pKa = 3.24VTEE57 pKa = 4.18THH59 pKa = 6.21WMFDD63 pKa = 3.88DD64 pKa = 4.13EE65 pKa = 4.87TMTDD69 pKa = 2.44AWTYY73 pKa = 9.1NGSVPGEE80 pKa = 4.15KK81 pKa = 9.33IRR83 pKa = 11.84VQEE86 pKa = 4.1GDD88 pKa = 3.71EE89 pKa = 4.66VILNVQNNLEE99 pKa = 4.38EE100 pKa = 4.23PTALHH105 pKa = 5.94LHH107 pKa = 6.61GFPVPNAMDD116 pKa = 4.31GVPGVTQNAIMPGEE130 pKa = 4.16EE131 pKa = 3.66FTYY134 pKa = 10.14EE135 pKa = 3.97YY136 pKa = 10.65QADD139 pKa = 3.82VPGTYY144 pKa = 8.57WYY146 pKa = 10.21HH147 pKa = 5.06SHH149 pKa = 6.73QDD151 pKa = 2.59GSTQVDD157 pKa = 3.14QGLYY161 pKa = 10.4GVFIVEE167 pKa = 4.62PEE169 pKa = 4.0DD170 pKa = 3.52QEE172 pKa = 5.41SYY174 pKa = 11.46DD175 pKa = 3.62VDD177 pKa = 3.55EE178 pKa = 5.56VIAIDD183 pKa = 4.06EE184 pKa = 4.38FAPMDD189 pKa = 3.96MDD191 pKa = 4.45MEE193 pKa = 7.08DD194 pKa = 5.29DD195 pKa = 3.36MHH197 pKa = 8.27EE198 pKa = 4.79DD199 pKa = 3.4MDD201 pKa = 5.08HH202 pKa = 8.31SDD204 pKa = 3.79MEE206 pKa = 4.89NGDD209 pKa = 4.14MEE211 pKa = 4.92EE212 pKa = 4.12MDD214 pKa = 4.54HH215 pKa = 8.64ADD217 pKa = 4.13MMNEE221 pKa = 3.99MYY223 pKa = 9.59DD224 pKa = 3.35TMVINGQSSSQIEE237 pKa = 4.37SVDD240 pKa = 3.44VEE242 pKa = 4.34EE243 pKa = 4.61GDD245 pKa = 3.73KK246 pKa = 11.32VKK248 pKa = 10.85LRR250 pKa = 11.84FVNAGLFTQIVSIPEE265 pKa = 3.65HH266 pKa = 6.03AFKK269 pKa = 9.94VTHH272 pKa = 6.73YY273 pKa = 10.59DD274 pKa = 3.45GQPVNEE280 pKa = 4.8PEE282 pKa = 4.46MISDD286 pKa = 3.83TSFQIAPAEE295 pKa = 4.15RR296 pKa = 11.84YY297 pKa = 9.59DD298 pKa = 4.67VEE300 pKa = 5.22IEE302 pKa = 3.91MDD304 pKa = 4.14NPGAWGIQVFAEE316 pKa = 4.19EE317 pKa = 4.0NQEE320 pKa = 4.0RR321 pKa = 11.84LNASLPLIYY330 pKa = 10.23DD331 pKa = 4.2GYY333 pKa = 10.98EE334 pKa = 4.26DD335 pKa = 5.38EE336 pKa = 5.89DD337 pKa = 4.45LQTGDD342 pKa = 4.27TPSSSLDD349 pKa = 3.07LTTYY353 pKa = 11.27GEE355 pKa = 4.18AQNMNEE361 pKa = 3.45YY362 pKa = 10.44DD363 pKa = 3.39INKK366 pKa = 9.43EE367 pKa = 3.42YY368 pKa = 11.4DD369 pKa = 3.47MNLGTDD375 pKa = 3.63DD376 pKa = 4.69GGDD379 pKa = 3.15TFTINGKK386 pKa = 7.75QFPDD390 pKa = 3.28HH391 pKa = 6.85EE392 pKa = 4.52IYY394 pKa = 10.37EE395 pKa = 4.27VEE397 pKa = 4.09EE398 pKa = 4.96GDD400 pKa = 3.97VVKK403 pKa = 9.71VTIEE407 pKa = 3.75NDD409 pKa = 3.51TEE411 pKa = 4.64DD412 pKa = 4.02DD413 pKa = 4.08HH414 pKa = 8.08PMHH417 pKa = 6.57LHH419 pKa = 6.7GEE421 pKa = 4.2FFDD424 pKa = 4.8VISKK428 pKa = 10.68DD429 pKa = 3.62GEE431 pKa = 4.14PLQGSSVTKK440 pKa = 9.47DD441 pKa = 3.32TLNVRR446 pKa = 11.84PGEE449 pKa = 4.02TYY451 pKa = 10.28EE452 pKa = 4.94IVFEE456 pKa = 4.22AQNPGNWMFHH466 pKa = 4.92CHH468 pKa = 5.55EE469 pKa = 4.51FHH471 pKa = 6.84HH472 pKa = 6.93ASDD475 pKa = 3.7GMVAEE480 pKa = 4.21VQYY483 pKa = 11.21EE484 pKa = 4.23GFEE487 pKa = 4.18PAFTPDD493 pKa = 3.86PNVPNQPEE501 pKa = 3.67
MM1 pKa = 7.17KK2 pKa = 9.99KK3 pKa = 10.46VYY5 pKa = 10.19IGMSTIFFVLAGCSGEE21 pKa = 4.2GLDD24 pKa = 4.5EE25 pKa = 4.86GSTGSTEE32 pKa = 4.08SQQEE36 pKa = 4.25GKK38 pKa = 10.15GNEE41 pKa = 3.94MMTEE45 pKa = 3.91DD46 pKa = 3.5GKK48 pKa = 11.21EE49 pKa = 3.96VFNLDD54 pKa = 3.24VTEE57 pKa = 4.18THH59 pKa = 6.21WMFDD63 pKa = 3.88DD64 pKa = 4.13EE65 pKa = 4.87TMTDD69 pKa = 2.44AWTYY73 pKa = 9.1NGSVPGEE80 pKa = 4.15KK81 pKa = 9.33IRR83 pKa = 11.84VQEE86 pKa = 4.1GDD88 pKa = 3.71EE89 pKa = 4.66VILNVQNNLEE99 pKa = 4.38EE100 pKa = 4.23PTALHH105 pKa = 5.94LHH107 pKa = 6.61GFPVPNAMDD116 pKa = 4.31GVPGVTQNAIMPGEE130 pKa = 4.16EE131 pKa = 3.66FTYY134 pKa = 10.14EE135 pKa = 3.97YY136 pKa = 10.65QADD139 pKa = 3.82VPGTYY144 pKa = 8.57WYY146 pKa = 10.21HH147 pKa = 5.06SHH149 pKa = 6.73QDD151 pKa = 2.59GSTQVDD157 pKa = 3.14QGLYY161 pKa = 10.4GVFIVEE167 pKa = 4.62PEE169 pKa = 4.0DD170 pKa = 3.52QEE172 pKa = 5.41SYY174 pKa = 11.46DD175 pKa = 3.62VDD177 pKa = 3.55EE178 pKa = 5.56VIAIDD183 pKa = 4.06EE184 pKa = 4.38FAPMDD189 pKa = 3.96MDD191 pKa = 4.45MEE193 pKa = 7.08DD194 pKa = 5.29DD195 pKa = 3.36MHH197 pKa = 8.27EE198 pKa = 4.79DD199 pKa = 3.4MDD201 pKa = 5.08HH202 pKa = 8.31SDD204 pKa = 3.79MEE206 pKa = 4.89NGDD209 pKa = 4.14MEE211 pKa = 4.92EE212 pKa = 4.12MDD214 pKa = 4.54HH215 pKa = 8.64ADD217 pKa = 4.13MMNEE221 pKa = 3.99MYY223 pKa = 9.59DD224 pKa = 3.35TMVINGQSSSQIEE237 pKa = 4.37SVDD240 pKa = 3.44VEE242 pKa = 4.34EE243 pKa = 4.61GDD245 pKa = 3.73KK246 pKa = 11.32VKK248 pKa = 10.85LRR250 pKa = 11.84FVNAGLFTQIVSIPEE265 pKa = 3.65HH266 pKa = 6.03AFKK269 pKa = 9.94VTHH272 pKa = 6.73YY273 pKa = 10.59DD274 pKa = 3.45GQPVNEE280 pKa = 4.8PEE282 pKa = 4.46MISDD286 pKa = 3.83TSFQIAPAEE295 pKa = 4.15RR296 pKa = 11.84YY297 pKa = 9.59DD298 pKa = 4.67VEE300 pKa = 5.22IEE302 pKa = 3.91MDD304 pKa = 4.14NPGAWGIQVFAEE316 pKa = 4.19EE317 pKa = 4.0NQEE320 pKa = 4.0RR321 pKa = 11.84LNASLPLIYY330 pKa = 10.23DD331 pKa = 4.2GYY333 pKa = 10.98EE334 pKa = 4.26DD335 pKa = 5.38EE336 pKa = 5.89DD337 pKa = 4.45LQTGDD342 pKa = 4.27TPSSSLDD349 pKa = 3.07LTTYY353 pKa = 11.27GEE355 pKa = 4.18AQNMNEE361 pKa = 3.45YY362 pKa = 10.44DD363 pKa = 3.39INKK366 pKa = 9.43EE367 pKa = 3.42YY368 pKa = 11.4DD369 pKa = 3.47MNLGTDD375 pKa = 3.63DD376 pKa = 4.69GGDD379 pKa = 3.15TFTINGKK386 pKa = 7.75QFPDD390 pKa = 3.28HH391 pKa = 6.85EE392 pKa = 4.52IYY394 pKa = 10.37EE395 pKa = 4.27VEE397 pKa = 4.09EE398 pKa = 4.96GDD400 pKa = 3.97VVKK403 pKa = 9.71VTIEE407 pKa = 3.75NDD409 pKa = 3.51TEE411 pKa = 4.64DD412 pKa = 4.02DD413 pKa = 4.08HH414 pKa = 8.08PMHH417 pKa = 6.57LHH419 pKa = 6.7GEE421 pKa = 4.2FFDD424 pKa = 4.8VISKK428 pKa = 10.68DD429 pKa = 3.62GEE431 pKa = 4.14PLQGSSVTKK440 pKa = 9.47DD441 pKa = 3.32TLNVRR446 pKa = 11.84PGEE449 pKa = 4.02TYY451 pKa = 10.28EE452 pKa = 4.94IVFEE456 pKa = 4.22AQNPGNWMFHH466 pKa = 4.92CHH468 pKa = 5.55EE469 pKa = 4.51FHH471 pKa = 6.84HH472 pKa = 6.93ASDD475 pKa = 3.7GMVAEE480 pKa = 4.21VQYY483 pKa = 11.21EE484 pKa = 4.23GFEE487 pKa = 4.18PAFTPDD493 pKa = 3.86PNVPNQPEE501 pKa = 3.67
Molecular weight: 56.33 kDa
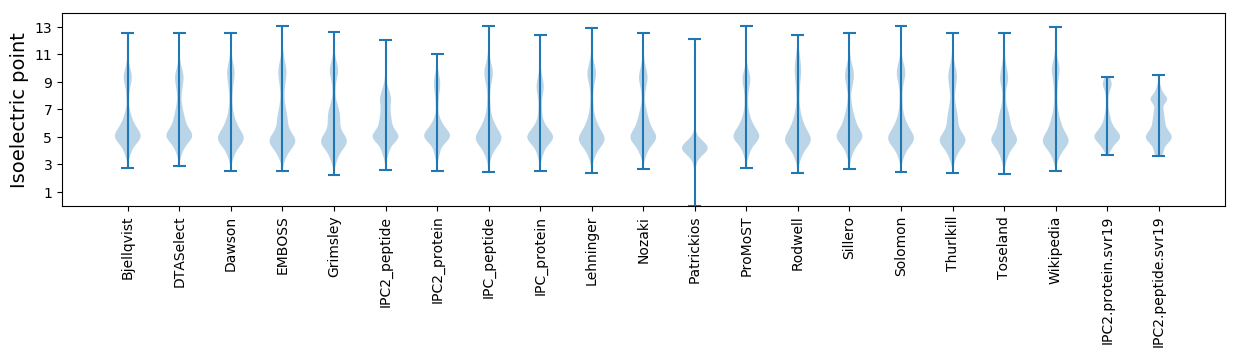
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BZX9|A0A345BZX9_9BACI Pyridoxal 5'-phosphate synthase subunit PdxS OS=Salicibibacter kimchii OX=2099786 GN=pdxS PE=3 SV=1
MM1 pKa = 7.7AKK3 pKa = 7.99MTFQPNNRR11 pKa = 11.84KK12 pKa = 9.31RR13 pKa = 11.84KK14 pKa = 8.54KK15 pKa = 8.5EE16 pKa = 3.58HH17 pKa = 5.81GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.28NGRR29 pKa = 11.84QVLKK33 pKa = 10.51RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.43KK38 pKa = 10.24GRR40 pKa = 11.84KK41 pKa = 8.58NLSAA45 pKa = 4.67
MM1 pKa = 7.7AKK3 pKa = 7.99MTFQPNNRR11 pKa = 11.84KK12 pKa = 9.31RR13 pKa = 11.84KK14 pKa = 8.54KK15 pKa = 8.5EE16 pKa = 3.58HH17 pKa = 5.81GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.28NGRR29 pKa = 11.84QVLKK33 pKa = 10.51RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.43KK38 pKa = 10.24GRR40 pKa = 11.84KK41 pKa = 8.58NLSAA45 pKa = 4.67
Molecular weight: 5.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
988862 |
26 |
1432 |
281.8 |
31.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.84 ± 0.039 | 0.651 ± 0.01 |
5.849 ± 0.042 | 8.184 ± 0.065 |
4.22 ± 0.035 | 7.253 ± 0.042 |
2.337 ± 0.018 | 6.86 ± 0.045 |
5.289 ± 0.041 | 9.329 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.038 ± 0.019 | 3.987 ± 0.025 |
3.862 ± 0.022 | 3.833 ± 0.032 |
4.731 ± 0.037 | 5.704 ± 0.029 |
5.574 ± 0.025 | 7.145 ± 0.033 |
1.048 ± 0.015 | 3.265 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
