
Brevundimonas sp. M20
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Brevundimonas; unclassified Brevundimonas
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
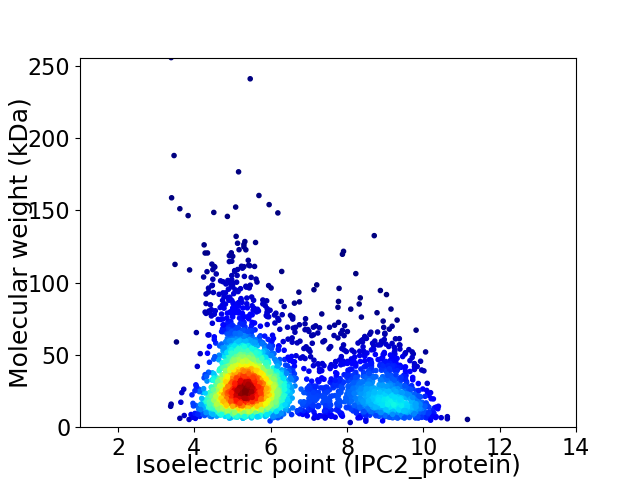
Virtual 2D-PAGE plot for 3160 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A514C4J0|A0A514C4J0_9CAUL Phosphonates import ATP-binding protein PhnC OS=Brevundimonas sp. M20 OX=2591463 GN=phnC PE=3 SV=1
MM1 pKa = 7.76SDD3 pKa = 2.9RR4 pKa = 11.84TAALPAGTALPPTLPLMADD23 pKa = 3.48DD24 pKa = 5.24RR25 pKa = 11.84FAPGITEE32 pKa = 4.07TGDD35 pKa = 3.22AGDD38 pKa = 3.47NFLYY42 pKa = 8.91GTHH45 pKa = 6.97NDD47 pKa = 3.43DD48 pKa = 4.44TLNGMAGNDD57 pKa = 3.63VLNGEE62 pKa = 5.29DD63 pKa = 4.38GYY65 pKa = 11.75DD66 pKa = 3.4WLIGGAGNDD75 pKa = 3.69VFNGGDD81 pKa = 3.78GIDD84 pKa = 3.54TGDD87 pKa = 3.59YY88 pKa = 9.89SAAASGVTARR98 pKa = 11.84LDD100 pKa = 3.02IGRR103 pKa = 11.84ATNDD107 pKa = 2.78GDD109 pKa = 4.02GGVDD113 pKa = 2.87ILNSIEE119 pKa = 4.11RR120 pKa = 11.84LYY122 pKa = 11.43GSAFNDD128 pKa = 3.36VLIGDD133 pKa = 4.22GQDD136 pKa = 3.11NLLVGGAGYY145 pKa = 10.74DD146 pKa = 3.63VLIGGAGNDD155 pKa = 3.61RR156 pKa = 11.84IYY158 pKa = 11.27GGVGAANEE166 pKa = 4.38LYY168 pKa = 10.73GGAGDD173 pKa = 3.73DD174 pKa = 4.48TFYY177 pKa = 11.61LDD179 pKa = 4.88ANDD182 pKa = 4.11TVVEE186 pKa = 4.06LAGGGIDD193 pKa = 3.51TVRR196 pKa = 11.84TSLTLVNLAANVEE209 pKa = 4.24NLWYY213 pKa = 10.1DD214 pKa = 4.01GPQDD218 pKa = 3.49FTGNGNAGDD227 pKa = 3.93NVITGGSGNDD237 pKa = 3.38VLRR240 pKa = 11.84GGVGNDD246 pKa = 3.35TLNGRR251 pKa = 11.84GGMDD255 pKa = 3.02TADD258 pKa = 3.8YY259 pKa = 8.91STAAAGIHH267 pKa = 6.23ARR269 pKa = 11.84LDD271 pKa = 3.34YY272 pKa = 8.15MTAFNDD278 pKa = 3.44GDD280 pKa = 4.21GGVDD284 pKa = 3.17TYY286 pKa = 11.97VDD288 pKa = 3.23IEE290 pKa = 4.94AIVGSAFDD298 pKa = 4.22DD299 pKa = 4.22LLVGGALNDD308 pKa = 3.96HH309 pKa = 6.81LSGGLGRR316 pKa = 11.84DD317 pKa = 3.4TLLGGTGNDD326 pKa = 3.53VLSGGQGVANTLQGGVGDD344 pKa = 4.75DD345 pKa = 4.91RR346 pKa = 11.84YY347 pKa = 11.03ILDD350 pKa = 4.06ANDD353 pKa = 4.33TIVEE357 pKa = 4.07LAGEE361 pKa = 4.45GFDD364 pKa = 3.47TVEE367 pKa = 3.97VHH369 pKa = 6.16IGAYY373 pKa = 10.07VMAANLEE380 pKa = 3.94NMEE383 pKa = 4.36YY384 pKa = 10.55VGQTTFNGTGNAGNNNIYY402 pKa = 10.77GGDD405 pKa = 3.77LNDD408 pKa = 3.46VLRR411 pKa = 11.84GGGGDD416 pKa = 3.31DD417 pKa = 3.24HH418 pKa = 8.34LYY420 pKa = 10.79GGRR423 pKa = 11.84GDD425 pKa = 3.69DD426 pKa = 3.59TVVLRR431 pKa = 11.84GAAGDD436 pKa = 3.75YY437 pKa = 9.53TITGDD442 pKa = 3.45GVGYY446 pKa = 10.19RR447 pKa = 11.84IVDD450 pKa = 3.93AVPGRR455 pKa = 11.84DD456 pKa = 2.8GSTYY460 pKa = 8.33VTGIEE465 pKa = 4.32TVLFEE470 pKa = 5.51ADD472 pKa = 3.02GTTRR476 pKa = 11.84TLSYY480 pKa = 9.6TPPPAPSEE488 pKa = 4.8LIDD491 pKa = 3.62KK492 pKa = 10.7ADD494 pKa = 3.7YY495 pKa = 11.4GDD497 pKa = 3.93AQVLPGVIDD506 pKa = 4.56DD507 pKa = 4.87AFLDD511 pKa = 3.7LGGVDD516 pKa = 5.93FGPQTLPPVFDD527 pKa = 5.11DD528 pKa = 6.15FLVLDD533 pKa = 5.55DD534 pKa = 5.0GLSDD538 pKa = 4.13PVWIAAAHH546 pKa = 5.53NAAPEE551 pKa = 4.01IQHH554 pKa = 6.62ILDD557 pKa = 4.01IFHH560 pKa = 7.26GDD562 pKa = 3.57DD563 pKa = 4.13AGHH566 pKa = 6.91HH567 pKa = 6.18GPFNAVDD574 pKa = 3.75PWAA577 pKa = 4.71
MM1 pKa = 7.76SDD3 pKa = 2.9RR4 pKa = 11.84TAALPAGTALPPTLPLMADD23 pKa = 3.48DD24 pKa = 5.24RR25 pKa = 11.84FAPGITEE32 pKa = 4.07TGDD35 pKa = 3.22AGDD38 pKa = 3.47NFLYY42 pKa = 8.91GTHH45 pKa = 6.97NDD47 pKa = 3.43DD48 pKa = 4.44TLNGMAGNDD57 pKa = 3.63VLNGEE62 pKa = 5.29DD63 pKa = 4.38GYY65 pKa = 11.75DD66 pKa = 3.4WLIGGAGNDD75 pKa = 3.69VFNGGDD81 pKa = 3.78GIDD84 pKa = 3.54TGDD87 pKa = 3.59YY88 pKa = 9.89SAAASGVTARR98 pKa = 11.84LDD100 pKa = 3.02IGRR103 pKa = 11.84ATNDD107 pKa = 2.78GDD109 pKa = 4.02GGVDD113 pKa = 2.87ILNSIEE119 pKa = 4.11RR120 pKa = 11.84LYY122 pKa = 11.43GSAFNDD128 pKa = 3.36VLIGDD133 pKa = 4.22GQDD136 pKa = 3.11NLLVGGAGYY145 pKa = 10.74DD146 pKa = 3.63VLIGGAGNDD155 pKa = 3.61RR156 pKa = 11.84IYY158 pKa = 11.27GGVGAANEE166 pKa = 4.38LYY168 pKa = 10.73GGAGDD173 pKa = 3.73DD174 pKa = 4.48TFYY177 pKa = 11.61LDD179 pKa = 4.88ANDD182 pKa = 4.11TVVEE186 pKa = 4.06LAGGGIDD193 pKa = 3.51TVRR196 pKa = 11.84TSLTLVNLAANVEE209 pKa = 4.24NLWYY213 pKa = 10.1DD214 pKa = 4.01GPQDD218 pKa = 3.49FTGNGNAGDD227 pKa = 3.93NVITGGSGNDD237 pKa = 3.38VLRR240 pKa = 11.84GGVGNDD246 pKa = 3.35TLNGRR251 pKa = 11.84GGMDD255 pKa = 3.02TADD258 pKa = 3.8YY259 pKa = 8.91STAAAGIHH267 pKa = 6.23ARR269 pKa = 11.84LDD271 pKa = 3.34YY272 pKa = 8.15MTAFNDD278 pKa = 3.44GDD280 pKa = 4.21GGVDD284 pKa = 3.17TYY286 pKa = 11.97VDD288 pKa = 3.23IEE290 pKa = 4.94AIVGSAFDD298 pKa = 4.22DD299 pKa = 4.22LLVGGALNDD308 pKa = 3.96HH309 pKa = 6.81LSGGLGRR316 pKa = 11.84DD317 pKa = 3.4TLLGGTGNDD326 pKa = 3.53VLSGGQGVANTLQGGVGDD344 pKa = 4.75DD345 pKa = 4.91RR346 pKa = 11.84YY347 pKa = 11.03ILDD350 pKa = 4.06ANDD353 pKa = 4.33TIVEE357 pKa = 4.07LAGEE361 pKa = 4.45GFDD364 pKa = 3.47TVEE367 pKa = 3.97VHH369 pKa = 6.16IGAYY373 pKa = 10.07VMAANLEE380 pKa = 3.94NMEE383 pKa = 4.36YY384 pKa = 10.55VGQTTFNGTGNAGNNNIYY402 pKa = 10.77GGDD405 pKa = 3.77LNDD408 pKa = 3.46VLRR411 pKa = 11.84GGGGDD416 pKa = 3.31DD417 pKa = 3.24HH418 pKa = 8.34LYY420 pKa = 10.79GGRR423 pKa = 11.84GDD425 pKa = 3.69DD426 pKa = 3.59TVVLRR431 pKa = 11.84GAAGDD436 pKa = 3.75YY437 pKa = 9.53TITGDD442 pKa = 3.45GVGYY446 pKa = 10.19RR447 pKa = 11.84IVDD450 pKa = 3.93AVPGRR455 pKa = 11.84DD456 pKa = 2.8GSTYY460 pKa = 8.33VTGIEE465 pKa = 4.32TVLFEE470 pKa = 5.51ADD472 pKa = 3.02GTTRR476 pKa = 11.84TLSYY480 pKa = 9.6TPPPAPSEE488 pKa = 4.8LIDD491 pKa = 3.62KK492 pKa = 10.7ADD494 pKa = 3.7YY495 pKa = 11.4GDD497 pKa = 3.93AQVLPGVIDD506 pKa = 4.56DD507 pKa = 4.87AFLDD511 pKa = 3.7LGGVDD516 pKa = 5.93FGPQTLPPVFDD527 pKa = 5.11DD528 pKa = 6.15FLVLDD533 pKa = 5.55DD534 pKa = 5.0GLSDD538 pKa = 4.13PVWIAAAHH546 pKa = 5.53NAAPEE551 pKa = 4.01IQHH554 pKa = 6.62ILDD557 pKa = 4.01IFHH560 pKa = 7.26GDD562 pKa = 3.57DD563 pKa = 4.13AGHH566 pKa = 6.91HH567 pKa = 6.18GPFNAVDD574 pKa = 3.75PWAA577 pKa = 4.71
Molecular weight: 58.91 kDa
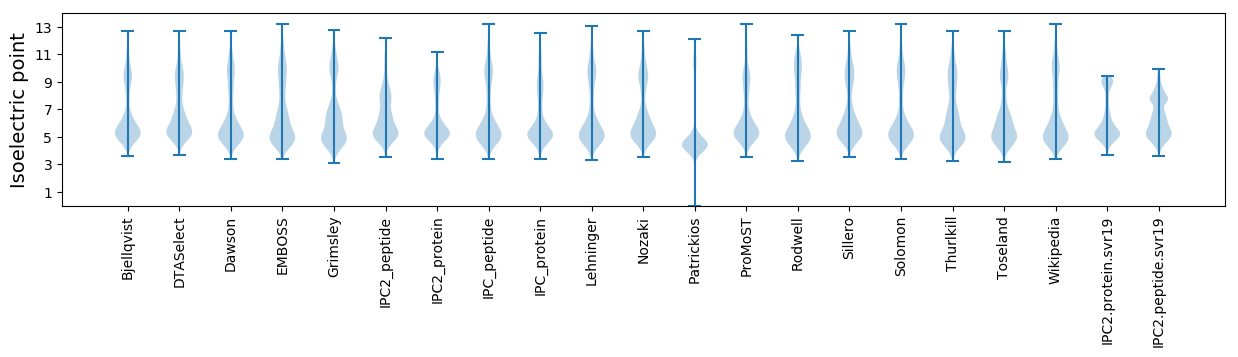
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A514BYB4|A0A514BYB4_9CAUL Uncharacterized protein OS=Brevundimonas sp. M20 OX=2591463 GN=FKQ52_02420 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.05NGQKK29 pKa = 9.47IVQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.05NGQKK29 pKa = 9.47IVQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
993302 |
30 |
2525 |
314.3 |
33.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.394 ± 0.06 | 0.696 ± 0.012 |
6.055 ± 0.038 | 5.712 ± 0.042 |
3.473 ± 0.023 | 9.146 ± 0.052 |
1.748 ± 0.022 | 4.455 ± 0.029 |
2.711 ± 0.042 | 9.85 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.02 | 2.435 ± 0.029 |
5.503 ± 0.035 | 3.095 ± 0.023 |
7.533 ± 0.049 | 5.11 ± 0.031 |
5.584 ± 0.04 | 7.657 ± 0.034 |
1.502 ± 0.017 | 2.053 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
