
Citrus leprosis virus C (isolate Citrus sinesis/Brazil/Cordeiropolis/2003) (CiLV-C)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Kitaviridae; Cilevirus; Citrus leprosis virus C
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
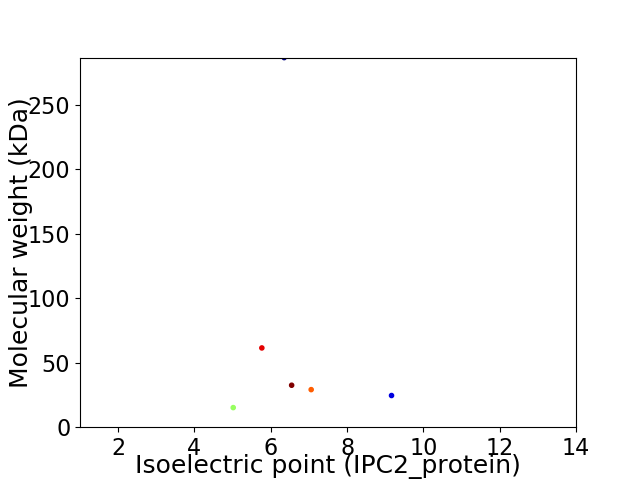
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
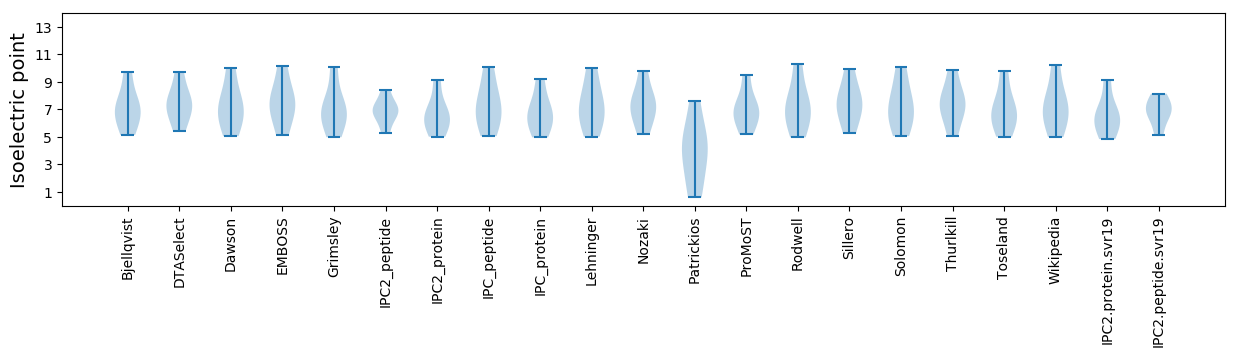
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q1KZ58|P29_CILVC P29 OS=Citrus leprosis virus C (isolate Citrus sinesis/Brazil/Cordeiropolis/2003) OX=686950 GN=p29 PE=4 SV=1
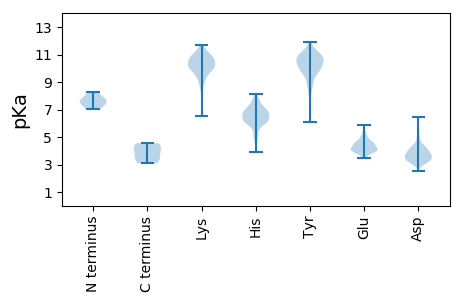
MM1 pKa = 7.4LNWSTIEE8 pKa = 4.21WDD10 pKa = 4.31SFWQQHH16 pKa = 6.14DD17 pKa = 3.98CGCFTFEE24 pKa = 4.92CDD26 pKa = 4.87FITSIDD32 pKa = 4.06PLVHH36 pKa = 7.38DD37 pKa = 3.78YY38 pKa = 11.34AIYY41 pKa = 10.49HH42 pKa = 6.08SLSQKK47 pKa = 9.03TVLEE51 pKa = 4.03MLQTHH56 pKa = 6.61LVAGPDD62 pKa = 3.11ASEE65 pKa = 4.08TIRR68 pKa = 11.84RR69 pKa = 11.84QVAFLIYY76 pKa = 10.14DD77 pKa = 3.6FHH79 pKa = 8.15RR80 pKa = 11.84LSCNCDD86 pKa = 2.78KK87 pKa = 11.24CYY89 pKa = 11.06GDD91 pKa = 5.22CNATTTGRR99 pKa = 11.84FKK101 pKa = 11.3VVDD104 pKa = 3.71RR105 pKa = 11.84VLNDD109 pKa = 3.93HH110 pKa = 6.84IEE112 pKa = 4.01FGIMRR117 pKa = 11.84RR118 pKa = 11.84QDD120 pKa = 4.79LIPILHH126 pKa = 6.24NLEE129 pKa = 4.15TT130 pKa = 4.13
MM1 pKa = 7.4LNWSTIEE8 pKa = 4.21WDD10 pKa = 4.31SFWQQHH16 pKa = 6.14DD17 pKa = 3.98CGCFTFEE24 pKa = 4.92CDD26 pKa = 4.87FITSIDD32 pKa = 4.06PLVHH36 pKa = 7.38DD37 pKa = 3.78YY38 pKa = 11.34AIYY41 pKa = 10.49HH42 pKa = 6.08SLSQKK47 pKa = 9.03TVLEE51 pKa = 4.03MLQTHH56 pKa = 6.61LVAGPDD62 pKa = 3.11ASEE65 pKa = 4.08TIRR68 pKa = 11.84RR69 pKa = 11.84QVAFLIYY76 pKa = 10.14DD77 pKa = 3.6FHH79 pKa = 8.15RR80 pKa = 11.84LSCNCDD86 pKa = 2.78KK87 pKa = 11.24CYY89 pKa = 11.06GDD91 pKa = 5.22CNATTTGRR99 pKa = 11.84FKK101 pKa = 11.3VVDD104 pKa = 3.71RR105 pKa = 11.84VLNDD109 pKa = 3.93HH110 pKa = 6.84IEE112 pKa = 4.01FGIMRR117 pKa = 11.84RR118 pKa = 11.84QDD120 pKa = 4.79LIPILHH126 pKa = 6.24NLEE129 pKa = 4.15TT130 pKa = 4.13
Molecular weight: 15.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q1KZ55|MVP_CILVC Putative movement protein OS=Citrus leprosis virus C (isolate Citrus sinesis/Brazil/Cordeiropolis/2003) OX=686950 GN=MP PE=3 SV=1
MM1 pKa = 8.27DD2 pKa = 4.88AQLLQANKK10 pKa = 10.18RR11 pKa = 11.84LLRR14 pKa = 11.84RR15 pKa = 11.84AANVRR20 pKa = 11.84QRR22 pKa = 11.84YY23 pKa = 9.26KK24 pKa = 10.31MLATEE29 pKa = 4.62SFVADD34 pKa = 3.82IKK36 pKa = 11.1QILLRR41 pKa = 11.84FIQKK45 pKa = 9.55PNVIIMYY52 pKa = 10.01ISVLVLFAAHH62 pKa = 6.95IDD64 pKa = 4.23SNTHH68 pKa = 7.67DD69 pKa = 4.24ILDD72 pKa = 4.23DD73 pKa = 4.2LAAQFPNNTFIEE85 pKa = 4.4WAKK88 pKa = 11.11SNFFRR93 pKa = 11.84ICGALVFIPVIIDD106 pKa = 3.29TEE108 pKa = 4.4EE109 pKa = 3.56KK110 pKa = 9.9HH111 pKa = 6.61RR112 pKa = 11.84NYY114 pKa = 10.28LALVIFVFLMGFPQRR129 pKa = 11.84SIMEE133 pKa = 3.88YY134 pKa = 9.83FIYY137 pKa = 10.5SISFHH142 pKa = 7.15VYY144 pKa = 9.71AKK146 pKa = 10.31AKK148 pKa = 10.45HH149 pKa = 5.03PVTRR153 pKa = 11.84IFIIGAAVFSCVMFGIFTNEE173 pKa = 3.88QLRR176 pKa = 11.84KK177 pKa = 9.98LYY179 pKa = 10.94AEE181 pKa = 4.3LPKK184 pKa = 10.77VPTHH188 pKa = 6.06PVAVNRR194 pKa = 11.84VEE196 pKa = 4.65KK197 pKa = 10.21VANRR201 pKa = 11.84ASRR204 pKa = 11.84VSTEE208 pKa = 3.74GTVNFGG214 pKa = 3.37
MM1 pKa = 8.27DD2 pKa = 4.88AQLLQANKK10 pKa = 10.18RR11 pKa = 11.84LLRR14 pKa = 11.84RR15 pKa = 11.84AANVRR20 pKa = 11.84QRR22 pKa = 11.84YY23 pKa = 9.26KK24 pKa = 10.31MLATEE29 pKa = 4.62SFVADD34 pKa = 3.82IKK36 pKa = 11.1QILLRR41 pKa = 11.84FIQKK45 pKa = 9.55PNVIIMYY52 pKa = 10.01ISVLVLFAAHH62 pKa = 6.95IDD64 pKa = 4.23SNTHH68 pKa = 7.67DD69 pKa = 4.24ILDD72 pKa = 4.23DD73 pKa = 4.2LAAQFPNNTFIEE85 pKa = 4.4WAKK88 pKa = 11.11SNFFRR93 pKa = 11.84ICGALVFIPVIIDD106 pKa = 3.29TEE108 pKa = 4.4EE109 pKa = 3.56KK110 pKa = 9.9HH111 pKa = 6.61RR112 pKa = 11.84NYY114 pKa = 10.28LALVIFVFLMGFPQRR129 pKa = 11.84SIMEE133 pKa = 3.88YY134 pKa = 9.83FIYY137 pKa = 10.5SISFHH142 pKa = 7.15VYY144 pKa = 9.71AKK146 pKa = 10.31AKK148 pKa = 10.45HH149 pKa = 5.03PVTRR153 pKa = 11.84IFIIGAAVFSCVMFGIFTNEE173 pKa = 3.88QLRR176 pKa = 11.84KK177 pKa = 9.98LYY179 pKa = 10.94AEE181 pKa = 4.3LPKK184 pKa = 10.77VPTHH188 pKa = 6.06PVAVNRR194 pKa = 11.84VEE196 pKa = 4.65KK197 pKa = 10.21VANRR201 pKa = 11.84ASRR204 pKa = 11.84VSTEE208 pKa = 3.74GTVNFGG214 pKa = 3.37
Molecular weight: 24.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3953 |
130 |
2512 |
658.8 |
74.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.034 ± 0.966 | 2.707 ± 0.522 |
6.704 ± 0.7 | 5.186 ± 0.239 |
5.667 ± 0.608 | 4.882 ± 0.511 |
2.757 ± 0.225 | 6.527 ± 0.599 |
6.046 ± 0.561 | 9.006 ± 0.431 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.998 ± 0.185 | 4.376 ± 0.462 |
3.693 ± 0.598 | 2.808 ± 0.924 |
5.515 ± 0.611 | 8.297 ± 0.785 |
5.237 ± 0.443 | 8.677 ± 0.534 |
0.582 ± 0.139 | 4.301 ± 0.799 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
