
Seonamhaeicola sp. S2-3
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Seonamhaeicola; unclassified Seonamhaeicola
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
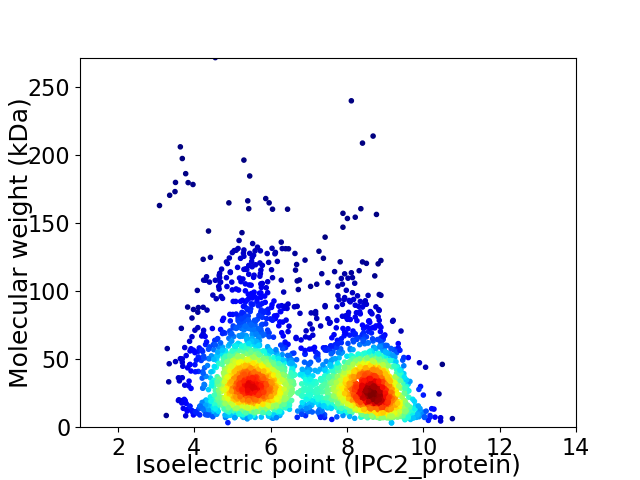
Virtual 2D-PAGE plot for 3101 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7DT73|A0A1U7DT73_9FLAO Cytochrome oxidase maturation protein cbb3-type OS=Seonamhaeicola sp. S2-3 OX=1936081 GN=BWZ22_10905 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 10.03IFFIGTIIMFFMFSCSDD19 pKa = 4.05DD20 pKa = 3.28IKK22 pKa = 10.77SYY24 pKa = 10.77PVGSDD29 pKa = 4.55FIDD32 pKa = 3.44NDD34 pKa = 3.62VNLKK38 pKa = 10.47VLDD41 pKa = 3.99TFTINAGTFKK51 pKa = 10.63LDD53 pKa = 3.43SLITSSTSRR62 pKa = 11.84ILLGSVKK69 pKa = 10.5DD70 pKa = 4.27DD71 pKa = 3.42YY72 pKa = 11.46FGNLTAQSYY81 pKa = 7.68FQVVNSDD88 pKa = 3.61FSISSDD94 pKa = 3.11AVYY97 pKa = 10.93DD98 pKa = 4.31SIGLVLNYY106 pKa = 10.68DD107 pKa = 3.24NYY109 pKa = 11.31YY110 pKa = 10.7YY111 pKa = 11.4GDD113 pKa = 3.51TTRR116 pKa = 11.84VQTYY120 pKa = 9.11KK121 pKa = 9.88VHH123 pKa = 7.08RR124 pKa = 11.84LLEE127 pKa = 4.28YY128 pKa = 10.64FEE130 pKa = 5.13PPEE133 pKa = 6.07DD134 pKa = 4.95DD135 pKa = 3.67DD136 pKa = 5.43AFYY139 pKa = 11.25NVSKK143 pKa = 11.04LEE145 pKa = 3.9YY146 pKa = 10.24DD147 pKa = 3.79EE148 pKa = 5.69EE149 pKa = 4.48PLGEE153 pKa = 4.36FTFSPRR159 pKa = 11.84PTSTSDD165 pKa = 3.32SIYY168 pKa = 10.69IPIDD172 pKa = 3.36KK173 pKa = 10.74DD174 pKa = 3.27LGEE177 pKa = 4.64EE178 pKa = 3.94IFNKK182 pKa = 9.75IRR184 pKa = 11.84DD185 pKa = 3.64NDD187 pKa = 3.69INSTDD192 pKa = 4.42DD193 pKa = 4.25FIQYY197 pKa = 10.12FYY199 pKa = 10.93GLTVIPDD206 pKa = 3.73TLVDD210 pKa = 4.09SNVLGFTYY218 pKa = 10.21TSSSDD223 pKa = 3.19IEE225 pKa = 4.58NNSGMRR231 pKa = 11.84IFYY234 pKa = 10.18TEE236 pKa = 5.25DD237 pKa = 3.35NDD239 pKa = 5.29DD240 pKa = 3.81TNEE243 pKa = 4.55DD244 pKa = 3.56NDD246 pKa = 4.07HH247 pKa = 6.67VINFYY252 pKa = 10.62IPSSDD257 pKa = 3.35KK258 pKa = 10.89QFNTIKK264 pKa = 10.62NNLSGTVIEE273 pKa = 4.91NLDD276 pKa = 3.82DD277 pKa = 3.81QEE279 pKa = 4.47TTIDD283 pKa = 5.17SYY285 pKa = 11.68DD286 pKa = 3.43TDD288 pKa = 3.42QLIFAQGGIGVSARR302 pKa = 11.84IEE304 pKa = 4.23MPTLKK309 pKa = 10.39YY310 pKa = 10.77LKK312 pKa = 9.04TLSDD316 pKa = 3.61EE317 pKa = 4.4STSLSAEE324 pKa = 4.02LTFAPLLNSYY334 pKa = 11.16DD335 pKa = 3.68EE336 pKa = 4.3TRR338 pKa = 11.84PLQDD342 pKa = 2.89SLAVYY347 pKa = 10.14IVDD350 pKa = 3.34NKK352 pKa = 10.5NRR354 pKa = 11.84IVSQLTDD361 pKa = 3.19ADD363 pKa = 4.4ANTSYY368 pKa = 11.39AILNQDD374 pKa = 2.44SDD376 pKa = 4.18EE377 pKa = 4.67FNEE380 pKa = 4.14STYY383 pKa = 11.37YY384 pKa = 10.64SIDD387 pKa = 2.97MSGFVEE393 pKa = 4.82TIFNSEE399 pKa = 3.48TDD401 pKa = 3.5LNYY404 pKa = 10.89AIMIQFIDD412 pKa = 3.59YY413 pKa = 10.15NKK415 pKa = 9.72TVDD418 pKa = 4.17SVIIDD423 pKa = 4.58DD424 pKa = 5.8LNEE427 pKa = 4.58DD428 pKa = 4.13DD429 pKa = 5.72NNIKK433 pKa = 10.79LSVTYY438 pKa = 10.53LEE440 pKa = 4.62YY441 pKa = 11.1
MM1 pKa = 7.8KK2 pKa = 10.03IFFIGTIIMFFMFSCSDD19 pKa = 4.05DD20 pKa = 3.28IKK22 pKa = 10.77SYY24 pKa = 10.77PVGSDD29 pKa = 4.55FIDD32 pKa = 3.44NDD34 pKa = 3.62VNLKK38 pKa = 10.47VLDD41 pKa = 3.99TFTINAGTFKK51 pKa = 10.63LDD53 pKa = 3.43SLITSSTSRR62 pKa = 11.84ILLGSVKK69 pKa = 10.5DD70 pKa = 4.27DD71 pKa = 3.42YY72 pKa = 11.46FGNLTAQSYY81 pKa = 7.68FQVVNSDD88 pKa = 3.61FSISSDD94 pKa = 3.11AVYY97 pKa = 10.93DD98 pKa = 4.31SIGLVLNYY106 pKa = 10.68DD107 pKa = 3.24NYY109 pKa = 11.31YY110 pKa = 10.7YY111 pKa = 11.4GDD113 pKa = 3.51TTRR116 pKa = 11.84VQTYY120 pKa = 9.11KK121 pKa = 9.88VHH123 pKa = 7.08RR124 pKa = 11.84LLEE127 pKa = 4.28YY128 pKa = 10.64FEE130 pKa = 5.13PPEE133 pKa = 6.07DD134 pKa = 4.95DD135 pKa = 3.67DD136 pKa = 5.43AFYY139 pKa = 11.25NVSKK143 pKa = 11.04LEE145 pKa = 3.9YY146 pKa = 10.24DD147 pKa = 3.79EE148 pKa = 5.69EE149 pKa = 4.48PLGEE153 pKa = 4.36FTFSPRR159 pKa = 11.84PTSTSDD165 pKa = 3.32SIYY168 pKa = 10.69IPIDD172 pKa = 3.36KK173 pKa = 10.74DD174 pKa = 3.27LGEE177 pKa = 4.64EE178 pKa = 3.94IFNKK182 pKa = 9.75IRR184 pKa = 11.84DD185 pKa = 3.64NDD187 pKa = 3.69INSTDD192 pKa = 4.42DD193 pKa = 4.25FIQYY197 pKa = 10.12FYY199 pKa = 10.93GLTVIPDD206 pKa = 3.73TLVDD210 pKa = 4.09SNVLGFTYY218 pKa = 10.21TSSSDD223 pKa = 3.19IEE225 pKa = 4.58NNSGMRR231 pKa = 11.84IFYY234 pKa = 10.18TEE236 pKa = 5.25DD237 pKa = 3.35NDD239 pKa = 5.29DD240 pKa = 3.81TNEE243 pKa = 4.55DD244 pKa = 3.56NDD246 pKa = 4.07HH247 pKa = 6.67VINFYY252 pKa = 10.62IPSSDD257 pKa = 3.35KK258 pKa = 10.89QFNTIKK264 pKa = 10.62NNLSGTVIEE273 pKa = 4.91NLDD276 pKa = 3.82DD277 pKa = 3.81QEE279 pKa = 4.47TTIDD283 pKa = 5.17SYY285 pKa = 11.68DD286 pKa = 3.43TDD288 pKa = 3.42QLIFAQGGIGVSARR302 pKa = 11.84IEE304 pKa = 4.23MPTLKK309 pKa = 10.39YY310 pKa = 10.77LKK312 pKa = 9.04TLSDD316 pKa = 3.61EE317 pKa = 4.4STSLSAEE324 pKa = 4.02LTFAPLLNSYY334 pKa = 11.16DD335 pKa = 3.68EE336 pKa = 4.3TRR338 pKa = 11.84PLQDD342 pKa = 2.89SLAVYY347 pKa = 10.14IVDD350 pKa = 3.34NKK352 pKa = 10.5NRR354 pKa = 11.84IVSQLTDD361 pKa = 3.19ADD363 pKa = 4.4ANTSYY368 pKa = 11.39AILNQDD374 pKa = 2.44SDD376 pKa = 4.18EE377 pKa = 4.67FNEE380 pKa = 4.14STYY383 pKa = 11.37YY384 pKa = 10.64SIDD387 pKa = 2.97MSGFVEE393 pKa = 4.82TIFNSEE399 pKa = 3.48TDD401 pKa = 3.5LNYY404 pKa = 10.89AIMIQFIDD412 pKa = 3.59YY413 pKa = 10.15NKK415 pKa = 9.72TVDD418 pKa = 4.17SVIIDD423 pKa = 4.58DD424 pKa = 5.8LNEE427 pKa = 4.58DD428 pKa = 4.13DD429 pKa = 5.72NNIKK433 pKa = 10.79LSVTYY438 pKa = 10.53LEE440 pKa = 4.62YY441 pKa = 11.1
Molecular weight: 50.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7DTE0|A0A1U7DTE0_9FLAO Uncharacterized protein OS=Seonamhaeicola sp. S2-3 OX=1936081 GN=BWZ22_10815 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.72LSVSSEE48 pKa = 3.76ARR50 pKa = 11.84HH51 pKa = 5.78KK52 pKa = 10.72KK53 pKa = 9.83
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.72LSVSSEE48 pKa = 3.76ARR50 pKa = 11.84HH51 pKa = 5.78KK52 pKa = 10.72KK53 pKa = 9.83
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1092078 |
26 |
2404 |
352.2 |
39.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.155 ± 0.039 | 0.762 ± 0.015 |
5.453 ± 0.035 | 6.464 ± 0.038 |
5.222 ± 0.035 | 6.14 ± 0.039 |
1.796 ± 0.02 | 8.078 ± 0.042 |
8.345 ± 0.055 | 9.251 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.009 ± 0.022 | 6.858 ± 0.05 |
3.337 ± 0.026 | 3.216 ± 0.023 |
3.186 ± 0.03 | 6.402 ± 0.037 |
5.88 ± 0.049 | 6.188 ± 0.033 |
1.046 ± 0.015 | 4.212 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
