
NL63-related bat coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Setracovirus; NL63-related bat coronavirus strain BtKYNL63-9b
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
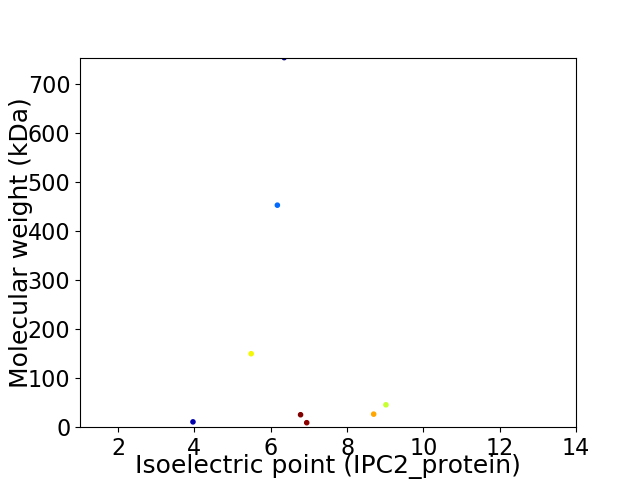
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
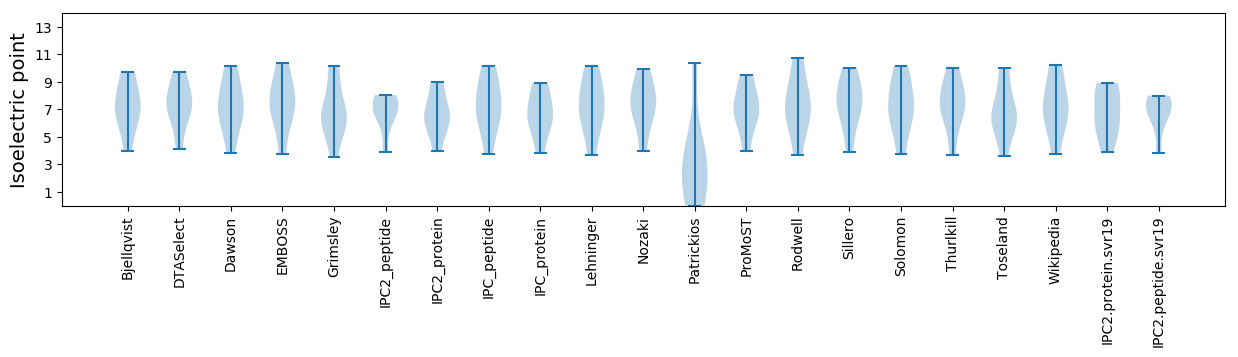
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L2KGB5|A0A1L2KGB5_9ALPC Isoform of A0A1L2KGB4 3C-like proteinase OS=NL63-related bat coronavirus OX=1920748 GN=orf1ab PE=3 SV=1
MM1 pKa = 8.03DD2 pKa = 5.11FMLFLYY8 pKa = 10.53ALSLFIEE15 pKa = 4.63EE16 pKa = 4.59GLPVAIALGVWAAEE30 pKa = 4.27VTGLVYY36 pKa = 10.2FLYY39 pKa = 10.76VDD41 pKa = 3.98GPVVLFTWNFLICYY55 pKa = 8.96AFLYY59 pKa = 9.91FILVPLVQEE68 pKa = 4.47LFLPGALDD76 pKa = 3.78LALDD80 pKa = 3.84QLRR83 pKa = 11.84GFHH86 pKa = 6.66DD87 pKa = 4.7FIVRR91 pKa = 11.84AMFCC95 pKa = 3.79
MM1 pKa = 8.03DD2 pKa = 5.11FMLFLYY8 pKa = 10.53ALSLFIEE15 pKa = 4.63EE16 pKa = 4.59GLPVAIALGVWAAEE30 pKa = 4.27VTGLVYY36 pKa = 10.2FLYY39 pKa = 10.76VDD41 pKa = 3.98GPVVLFTWNFLICYY55 pKa = 8.96AFLYY59 pKa = 9.91FILVPLVQEE68 pKa = 4.47LFLPGALDD76 pKa = 3.78LALDD80 pKa = 3.84QLRR83 pKa = 11.84GFHH86 pKa = 6.66DD87 pKa = 4.7FIVRR91 pKa = 11.84AMFCC95 pKa = 3.79
Molecular weight: 10.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L2KGC0|A0A1L2KGC0_9ALPC Membrane protein OS=NL63-related bat coronavirus OX=1920748 GN=M PE=3 SV=1
MM1 pKa = 7.26ATVNWADD8 pKa = 3.82DD9 pKa = 3.51KK10 pKa = 11.1RR11 pKa = 11.84GKK13 pKa = 9.97RR14 pKa = 11.84KK15 pKa = 9.77YY16 pKa = 9.36PPPSFYY22 pKa = 11.06LPLVVKK28 pKa = 10.43SDD30 pKa = 3.34KK31 pKa = 10.59QPYY34 pKa = 9.07KK35 pKa = 10.55VIPRR39 pKa = 11.84NLVPKK44 pKa = 10.77GKK46 pKa = 10.37GNKK49 pKa = 8.87DD50 pKa = 3.28QQIGYY55 pKa = 9.33WNVQRR60 pKa = 11.84RR61 pKa = 11.84WRR63 pKa = 11.84MRR65 pKa = 11.84KK66 pKa = 7.78GQRR69 pKa = 11.84VDD71 pKa = 3.01IDD73 pKa = 4.02PKK75 pKa = 9.33VHH77 pKa = 7.15FYY79 pKa = 11.42YY80 pKa = 10.81LGTGPRR86 pKa = 11.84ADD88 pKa = 3.08LKK90 pKa = 10.72FRR92 pKa = 11.84EE93 pKa = 4.26RR94 pKa = 11.84AEE96 pKa = 4.14DD97 pKa = 3.93VVWVAMQGSKK107 pKa = 9.31TEE109 pKa = 4.0PTNLGNRR116 pKa = 11.84KK117 pKa = 9.39RR118 pKa = 11.84NQKK121 pKa = 9.76PIQPEE126 pKa = 3.58FDD128 pKa = 2.91IQLPNEE134 pKa = 4.15LEE136 pKa = 4.17VVEE139 pKa = 5.57FEE141 pKa = 4.89DD142 pKa = 4.91RR143 pKa = 11.84SNSSSRR149 pKa = 11.84ASSRR153 pKa = 11.84ASSRR157 pKa = 11.84GNSRR161 pKa = 11.84EE162 pKa = 3.71TSRR165 pKa = 11.84SNSRR169 pKa = 11.84QQSRR173 pKa = 11.84DD174 pKa = 2.95NSRR177 pKa = 11.84SPSRR181 pKa = 11.84SRR183 pKa = 11.84SNSTSEE189 pKa = 3.96SSQNSAQDD197 pKa = 3.11LVAAVTAALKK207 pKa = 10.74NLGFEE212 pKa = 4.48PPKK215 pKa = 10.38SDD217 pKa = 3.67KK218 pKa = 11.12SGNASGTSTPKK229 pKa = 10.57GKK231 pKa = 10.35KK232 pKa = 9.32KK233 pKa = 10.24PKK235 pKa = 8.18QAKK238 pKa = 9.82SNEE241 pKa = 4.0QGSPNNVPSDD251 pKa = 3.34KK252 pKa = 11.25SQMNKK257 pKa = 9.89PKK259 pKa = 10.06WKK261 pKa = 9.61RR262 pKa = 11.84VPNASEE268 pKa = 4.32NVIKK272 pKa = 10.87CFGPRR277 pKa = 11.84DD278 pKa = 3.61FDD280 pKa = 5.02HH281 pKa = 7.2NMGDD285 pKa = 3.7ADD287 pKa = 4.08LVQNGVEE294 pKa = 4.3AKK296 pKa = 10.2NFPQIAEE303 pKa = 5.01LIPTQAAMFFDD314 pKa = 4.43SEE316 pKa = 4.57VSTKK320 pKa = 10.8EE321 pKa = 3.64MGNKK325 pKa = 8.67VQIIYY330 pKa = 8.58TYY332 pKa = 11.34KK333 pKa = 10.23MLVDD337 pKa = 4.6KK338 pKa = 10.89DD339 pKa = 3.9NKK341 pKa = 10.15HH342 pKa = 5.78LPKK345 pKa = 10.47FLEE348 pKa = 4.12QVSAFTKK355 pKa = 9.5PSVVKK360 pKa = 8.77EE361 pKa = 4.49TQSHH365 pKa = 6.79PLQNTMPEE373 pKa = 4.05PAQLNVAAAEE383 pKa = 4.14FKK385 pKa = 11.01PPVTTASDD393 pKa = 3.55GSNAEE398 pKa = 3.77IEE400 pKa = 4.33IVDD403 pKa = 4.18EE404 pKa = 4.24VLHH407 pKa = 6.31
MM1 pKa = 7.26ATVNWADD8 pKa = 3.82DD9 pKa = 3.51KK10 pKa = 11.1RR11 pKa = 11.84GKK13 pKa = 9.97RR14 pKa = 11.84KK15 pKa = 9.77YY16 pKa = 9.36PPPSFYY22 pKa = 11.06LPLVVKK28 pKa = 10.43SDD30 pKa = 3.34KK31 pKa = 10.59QPYY34 pKa = 9.07KK35 pKa = 10.55VIPRR39 pKa = 11.84NLVPKK44 pKa = 10.77GKK46 pKa = 10.37GNKK49 pKa = 8.87DD50 pKa = 3.28QQIGYY55 pKa = 9.33WNVQRR60 pKa = 11.84RR61 pKa = 11.84WRR63 pKa = 11.84MRR65 pKa = 11.84KK66 pKa = 7.78GQRR69 pKa = 11.84VDD71 pKa = 3.01IDD73 pKa = 4.02PKK75 pKa = 9.33VHH77 pKa = 7.15FYY79 pKa = 11.42YY80 pKa = 10.81LGTGPRR86 pKa = 11.84ADD88 pKa = 3.08LKK90 pKa = 10.72FRR92 pKa = 11.84EE93 pKa = 4.26RR94 pKa = 11.84AEE96 pKa = 4.14DD97 pKa = 3.93VVWVAMQGSKK107 pKa = 9.31TEE109 pKa = 4.0PTNLGNRR116 pKa = 11.84KK117 pKa = 9.39RR118 pKa = 11.84NQKK121 pKa = 9.76PIQPEE126 pKa = 3.58FDD128 pKa = 2.91IQLPNEE134 pKa = 4.15LEE136 pKa = 4.17VVEE139 pKa = 5.57FEE141 pKa = 4.89DD142 pKa = 4.91RR143 pKa = 11.84SNSSSRR149 pKa = 11.84ASSRR153 pKa = 11.84ASSRR157 pKa = 11.84GNSRR161 pKa = 11.84EE162 pKa = 3.71TSRR165 pKa = 11.84SNSRR169 pKa = 11.84QQSRR173 pKa = 11.84DD174 pKa = 2.95NSRR177 pKa = 11.84SPSRR181 pKa = 11.84SRR183 pKa = 11.84SNSTSEE189 pKa = 3.96SSQNSAQDD197 pKa = 3.11LVAAVTAALKK207 pKa = 10.74NLGFEE212 pKa = 4.48PPKK215 pKa = 10.38SDD217 pKa = 3.67KK218 pKa = 11.12SGNASGTSTPKK229 pKa = 10.57GKK231 pKa = 10.35KK232 pKa = 9.32KK233 pKa = 10.24PKK235 pKa = 8.18QAKK238 pKa = 9.82SNEE241 pKa = 4.0QGSPNNVPSDD251 pKa = 3.34KK252 pKa = 11.25SQMNKK257 pKa = 9.89PKK259 pKa = 10.06WKK261 pKa = 9.61RR262 pKa = 11.84VPNASEE268 pKa = 4.32NVIKK272 pKa = 10.87CFGPRR277 pKa = 11.84DD278 pKa = 3.61FDD280 pKa = 5.02HH281 pKa = 7.2NMGDD285 pKa = 3.7ADD287 pKa = 4.08LVQNGVEE294 pKa = 4.3AKK296 pKa = 10.2NFPQIAEE303 pKa = 5.01LIPTQAAMFFDD314 pKa = 4.43SEE316 pKa = 4.57VSTKK320 pKa = 10.8EE321 pKa = 3.64MGNKK325 pKa = 8.67VQIIYY330 pKa = 8.58TYY332 pKa = 11.34KK333 pKa = 10.23MLVDD337 pKa = 4.6KK338 pKa = 10.89DD339 pKa = 3.9NKK341 pKa = 10.15HH342 pKa = 5.78LPKK345 pKa = 10.47FLEE348 pKa = 4.12QVSAFTKK355 pKa = 9.5PSVVKK360 pKa = 8.77EE361 pKa = 4.49TQSHH365 pKa = 6.79PLQNTMPEE373 pKa = 4.05PAQLNVAAAEE383 pKa = 4.14FKK385 pKa = 11.01PPVTTASDD393 pKa = 3.55GSNAEE398 pKa = 3.77IEE400 pKa = 4.33IVDD403 pKa = 4.18EE404 pKa = 4.24VLHH407 pKa = 6.31
Molecular weight: 45.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13252 |
77 |
6758 |
1656.5 |
184.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.807 ± 0.247 | 3.192 ± 0.266 |
5.652 ± 0.335 | 3.894 ± 0.225 |
5.886 ± 0.35 | 6.58 ± 0.346 |
1.637 ± 0.104 | 4.694 ± 0.375 |
5.946 ± 0.494 | 9.417 ± 0.695 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.932 ± 0.189 | 5.516 ± 0.255 |
3.63 ± 0.223 | 2.92 ± 0.43 |
3.441 ± 0.205 | 7.516 ± 0.553 |
5.75 ± 0.411 | 9.931 ± 0.355 |
1.268 ± 0.193 | 4.377 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
