
Trichormus variabilis SAG 1403-4b
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Trichormus; Trichormus variabilis
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
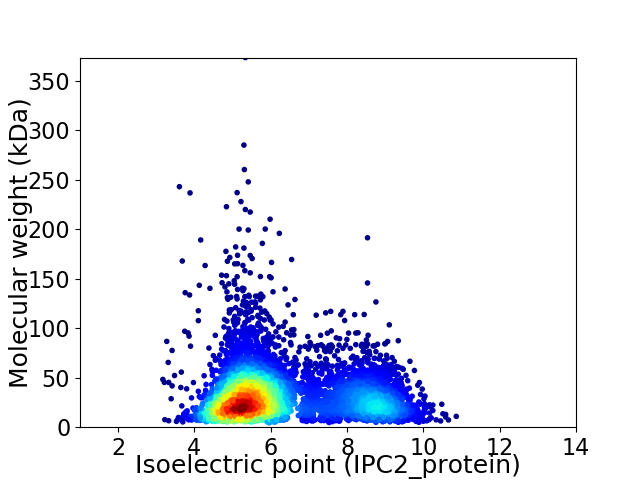
Virtual 2D-PAGE plot for 5168 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S5K2M9|A0A3S5K2M9_ANAVA DUF262 domain-containing protein OS=Trichormus variabilis SAG 1403-4b OX=447716 GN=DSM107003_49640 PE=4 SV=1
MM1 pKa = 7.51SRR3 pKa = 11.84PQVKK7 pKa = 8.45ITVSASGGPLNTPSYY22 pKa = 11.08KK23 pKa = 10.6DD24 pKa = 3.35LTITGIGTDD33 pKa = 3.27NTYY36 pKa = 9.32TFLNQQGVLDD46 pKa = 4.29AWCLDD51 pKa = 3.38SATGISSNTTYY62 pKa = 9.86TANVYY67 pKa = 10.52SSYY70 pKa = 11.25EE71 pKa = 3.96LGALATLGTPARR83 pKa = 11.84IGNLYY88 pKa = 10.75NLDD91 pKa = 3.66LVNWIINQNFAANPQYY107 pKa = 11.31NYY109 pKa = 11.33GEE111 pKa = 4.0IQAAIWTLLGDD122 pKa = 4.53TYY124 pKa = 11.37NSSGADD130 pKa = 3.34LTRR133 pKa = 11.84NGAVVASDD141 pKa = 3.64VNSIIALAKK150 pKa = 10.31ANGEE154 pKa = 4.04NFVPDD159 pKa = 3.71TNQFIGVIFDD169 pKa = 3.81PFVVNGDD176 pKa = 3.55GSVTPRR182 pKa = 11.84QPLIAQIKK190 pKa = 8.18AAKK193 pKa = 10.34LGDD196 pKa = 4.28LVWNDD201 pKa = 3.39LNANGIQDD209 pKa = 3.65TGEE212 pKa = 3.92SGIAGVTVNLTRR224 pKa = 11.84DD225 pKa = 3.52SNSNGIIEE233 pKa = 4.47SNEE236 pKa = 3.42ILATTTTNVSGLYY249 pKa = 9.12EE250 pKa = 4.26FRR252 pKa = 11.84GLTPGLSYY260 pKa = 10.35QVQFATPTGFDD271 pKa = 3.13AVSPRR276 pKa = 11.84QQGGNQTTDD285 pKa = 2.4SDD287 pKa = 4.27GLVSNVVILNPGEE300 pKa = 3.98FNQTIDD306 pKa = 3.17AGFYY310 pKa = 10.2KK311 pKa = 10.51YY312 pKa = 11.03ASLGDD317 pKa = 3.59FVFNDD322 pKa = 3.68GNNNGIQDD330 pKa = 3.76AGEE333 pKa = 4.05AGIGGVTVEE342 pKa = 5.58LINPANGSVIATTTTDD358 pKa = 3.82GIGAYY363 pKa = 8.44TFTGLTPGDD372 pKa = 3.82YY373 pKa = 9.53QVKK376 pKa = 8.04FTSPTGYY383 pKa = 10.33VFSTANVGDD392 pKa = 3.85DD393 pKa = 4.3TKK395 pKa = 11.6DD396 pKa = 3.11SDD398 pKa = 4.67ANTSTGLTKK407 pKa = 10.23TVTLTSGEE415 pKa = 4.31FNGTLDD421 pKa = 3.63AGLVQLASLGDD432 pKa = 3.93KK433 pKa = 10.5VFHH436 pKa = 7.25DD437 pKa = 5.01LDD439 pKa = 5.64ADD441 pKa = 5.05GIQDD445 pKa = 3.86AGEE448 pKa = 3.86QGIANASVKK457 pKa = 10.77LLDD460 pKa = 3.56TAGNVIATTTTDD472 pKa = 3.08GDD474 pKa = 3.85GLYY477 pKa = 10.64SFTNLQPGDD486 pKa = 3.73YY487 pKa = 9.59KK488 pKa = 11.37VQFVQPTGFNGVSLANVGGDD508 pKa = 4.01DD509 pKa = 4.78SVDD512 pKa = 3.57SDD514 pKa = 4.01GLISDD519 pKa = 4.49VVNLSPGEE527 pKa = 3.94NDD529 pKa = 3.38TTVDD533 pKa = 2.85AGFYY537 pKa = 8.57KK538 pKa = 10.21TASLGDD544 pKa = 3.5FVFNDD549 pKa = 3.68GNNNGIQDD557 pKa = 3.76AGEE560 pKa = 4.06AGIGEE565 pKa = 4.41VTVEE569 pKa = 5.02LINPADD575 pKa = 3.81GSVIATTTTDD585 pKa = 2.84EE586 pKa = 4.13TGAYY590 pKa = 7.47TFTGLTPGDD599 pKa = 3.82YY600 pKa = 9.53QVKK603 pKa = 8.25FTSPTDD609 pKa = 3.87YY610 pKa = 11.14VFSTADD616 pKa = 2.99QGGDD620 pKa = 3.44DD621 pKa = 4.5TVDD624 pKa = 3.2SDD626 pKa = 5.04ANTSTGLTQTVTLTSGEE643 pKa = 4.31FNGTLDD649 pKa = 3.63AGLVQLASLGDD660 pKa = 3.84FVFEE664 pKa = 4.86DD665 pKa = 3.94SNANGIQDD673 pKa = 3.09AWEE676 pKa = 3.81QGIAEE681 pKa = 4.27ASVNLLDD688 pKa = 3.78TAGNVIATTTTDD700 pKa = 3.08GDD702 pKa = 3.85GLYY705 pKa = 10.64SFTNLQPGDD714 pKa = 3.73YY715 pKa = 9.59KK716 pKa = 11.41VQFVQPAGFSGVSPSNVGGDD736 pKa = 3.97DD737 pKa = 4.02SVDD740 pKa = 3.57SDD742 pKa = 4.01GLISDD747 pKa = 4.49VVNLSPGEE755 pKa = 3.94NDD757 pKa = 3.38TTVDD761 pKa = 2.85AGFYY765 pKa = 8.57KK766 pKa = 10.21TASLGDD772 pKa = 3.5FVFNDD777 pKa = 3.68GNNNGIQDD785 pKa = 3.45IGEE788 pKa = 4.16AGIGGVTVEE797 pKa = 5.58LINPANGSVIASVITDD813 pKa = 3.89DD814 pKa = 3.48NGGYY818 pKa = 9.52QFSGLTPGDD827 pKa = 3.7YY828 pKa = 9.61QVKK831 pKa = 8.04FTSPTGYY838 pKa = 10.34VFSTAYY844 pKa = 10.15QGGDD848 pKa = 3.36DD849 pKa = 4.57TVDD852 pKa = 3.2SDD854 pKa = 5.04ANTSTGLTKK863 pKa = 10.23TVTLTSGEE871 pKa = 4.31FNGTLDD877 pKa = 3.63AGLVQLASLGDD888 pKa = 3.93KK889 pKa = 10.5VFHH892 pKa = 7.25DD893 pKa = 5.01LDD895 pKa = 5.64ADD897 pKa = 5.05GIQDD901 pKa = 3.86AGEE904 pKa = 3.86QGIANASVKK913 pKa = 10.77LLDD916 pKa = 3.56TAGNVIATTTTDD928 pKa = 3.08GDD930 pKa = 3.88GLYY933 pKa = 10.82SFINLQPGDD942 pKa = 3.83YY943 pKa = 9.82KK944 pKa = 11.37VQFVQPTDD952 pKa = 3.8FNGVSLANVGGDD964 pKa = 4.01DD965 pKa = 4.78SVDD968 pKa = 3.57SDD970 pKa = 4.01GLISDD975 pKa = 4.49VVNLSPGEE983 pKa = 3.94NDD985 pKa = 3.38TTVDD989 pKa = 2.85AGFYY993 pKa = 8.57KK994 pKa = 10.21TASLGDD1000 pKa = 3.5FVFNDD1005 pKa = 3.68GNNNGIQDD1013 pKa = 3.76AGEE1016 pKa = 4.05AGIGGVTVKK1025 pKa = 10.8LINPANGNVISTTTTDD1041 pKa = 3.03GAGAYY1046 pKa = 7.07TFTGLTPGDD1055 pKa = 3.84YY1056 pKa = 9.35QVKK1059 pKa = 9.7FISPTGYY1066 pKa = 10.67SFTAANQGGNDD1077 pKa = 4.28AVDD1080 pKa = 3.56SDD1082 pKa = 4.92ANPSTGLTQTVTLTSGEE1099 pKa = 4.32YY1100 pKa = 10.52NGTLDD1105 pKa = 4.27AGLVQLASLGDD1116 pKa = 3.93KK1117 pKa = 10.5VFHH1120 pKa = 7.25DD1121 pKa = 5.01LDD1123 pKa = 5.64ADD1125 pKa = 5.05GIQDD1129 pKa = 3.86AGEE1132 pKa = 3.86QGIANASVKK1141 pKa = 10.77LLDD1144 pKa = 3.56TAGNVIATTTTDD1156 pKa = 3.08GDD1158 pKa = 3.85GLYY1161 pKa = 10.64SFTNLQPGDD1170 pKa = 3.73YY1171 pKa = 9.59KK1172 pKa = 11.37VQFVQPTGFNGVSLANVGGDD1192 pKa = 3.78DD1193 pKa = 5.29SVDD1196 pKa = 3.18SDD1198 pKa = 3.89AASNLTTGVVNLSPGEE1214 pKa = 3.94NDD1216 pKa = 3.38TTVDD1220 pKa = 2.85AGFYY1224 pKa = 8.64KK1225 pKa = 10.25TASLGNFVFNDD1236 pKa = 3.34VNNNGIQEE1244 pKa = 3.98ASEE1247 pKa = 4.27AGVAGVTVTLTGGGADD1263 pKa = 3.84GLISTTADD1271 pKa = 2.81NTTITTTTNASGKK1284 pKa = 10.27YY1285 pKa = 9.23NFAGLTPGQEE1295 pKa = 4.26YY1296 pKa = 10.07QVGFSNLPAGYY1307 pKa = 10.36QFTQANAGTDD1317 pKa = 3.53DD1318 pKa = 5.35ALDD1321 pKa = 3.86SDD1323 pKa = 5.42ANTSTGKK1330 pKa = 7.77TQIVTLASGEE1340 pKa = 4.29NNTTLDD1346 pKa = 3.17AGIYY1350 pKa = 9.65QNAGDD1355 pKa = 4.69LSITKK1360 pKa = 9.53TNGLTTVSPGQQITYY1375 pKa = 9.2TIVAQNNGLITATNALVSDD1394 pKa = 5.22IIPSNLTNVTWTSVATGGATDD1415 pKa = 3.9NQSSGTGSINDD1426 pKa = 4.01YY1427 pKa = 9.5VTLTAGSSITYY1438 pKa = 8.23TVTGTVAPNALTPVYY1453 pKa = 8.18NTPTTINLTSNSSSALNGTAGNIRR1477 pKa = 11.84NFSANGISVNASAFSSTSTGTWNTAYY1503 pKa = 10.68LGLYY1507 pKa = 9.38GSYY1510 pKa = 11.06GLGVTDD1516 pKa = 4.27GGEE1519 pKa = 3.89NGSNGTHH1526 pKa = 7.02HH1527 pKa = 7.36IDD1529 pKa = 3.12NDD1531 pKa = 3.32GRR1533 pKa = 11.84NNYY1536 pKa = 9.11VLLEE1540 pKa = 4.14FSDD1543 pKa = 3.85SVVIDD1548 pKa = 3.95KK1549 pKa = 10.9AALSYY1554 pKa = 11.27VVGDD1558 pKa = 3.5SDD1560 pKa = 3.62ISVWIGNFNNPYY1572 pKa = 9.47TNHH1575 pKa = 6.17LTLNSSVLSSFGFTEE1590 pKa = 3.95INLGGSSDD1598 pKa = 3.79RR1599 pKa = 11.84NADD1602 pKa = 3.12INASNIVGNAVVIAAKK1618 pKa = 10.73VGDD1621 pKa = 3.98TNDD1624 pKa = 3.43EE1625 pKa = 4.37FKK1627 pKa = 11.13LKK1629 pKa = 10.81SLDD1632 pKa = 3.5IIKK1635 pKa = 9.23ATTQTGGSLVNTATITAPTGFTDD1658 pKa = 3.58TNSNNNSATDD1668 pKa = 3.23TDD1670 pKa = 4.38TIVASSTVRR1679 pKa = 11.84IGDD1682 pKa = 3.74RR1683 pKa = 11.84VWYY1686 pKa = 7.32DD1687 pKa = 3.32TNANGIQEE1695 pKa = 4.16TGEE1698 pKa = 4.0AGVAGVAVNLFNQAGTQVGTTTTDD1722 pKa = 2.83TNGFYY1727 pKa = 10.54QFTANTNSTYY1737 pKa = 10.71SVQFVQPTGFSGFTTANVGNDD1758 pKa = 3.61GADD1761 pKa = 3.24SDD1763 pKa = 4.55VVNANGTTAQFLVGTTDD1780 pKa = 3.26NLTIDD1785 pKa = 3.64AGLIKK1790 pKa = 10.76NIGDD1794 pKa = 3.81LSITKK1799 pKa = 9.48TDD1801 pKa = 3.16GLTTVTAGQQLTYY1814 pKa = 10.31TIVVSNNGFITATNALVTDD1833 pKa = 5.92AIPALLTNVTWTSVASGGATGNEE1856 pKa = 3.93TSGTGNINDD1865 pKa = 3.9YY1866 pKa = 9.57VTLTGGSSITYY1877 pKa = 7.49TVNGTVAANAVSSSSTYY1894 pKa = 9.84TDD1896 pKa = 2.52TRR1898 pKa = 11.84FDD1900 pKa = 4.14FNGNSSSDD1908 pKa = 3.44GTDD1911 pKa = 2.82GNTRR1915 pKa = 11.84TFTKK1919 pKa = 10.61DD1920 pKa = 2.88GITVTARR1927 pKa = 11.84AFSRR1931 pKa = 11.84VDD1933 pKa = 3.26GTNGAWSKK1941 pKa = 11.29AYY1943 pKa = 10.28LGSYY1947 pKa = 9.13TGGLGVTDD1955 pKa = 4.03SSEE1958 pKa = 3.83SSSYY1962 pKa = 10.79HH1963 pKa = 6.01RR1964 pKa = 11.84VDD1966 pKa = 3.43NGGGRR1971 pKa = 11.84DD1972 pKa = 3.58NYY1974 pKa = 10.9VLFQFSEE1981 pKa = 4.37AVVVDD1986 pKa = 3.89SAYY1989 pKa = 10.66LQYY1992 pKa = 11.4VVTDD1996 pKa = 3.36SDD1998 pKa = 3.82TSVWIGNFNNTITTLSDD2015 pKa = 3.22SVLNSFGFTEE2025 pKa = 4.4VNLGSGSDD2033 pKa = 3.11RR2034 pKa = 11.84WADD2037 pKa = 3.27INAGNYY2043 pKa = 9.46LGNTLVIAAKK2053 pKa = 9.9TDD2055 pKa = 3.43DD2056 pKa = 3.81TNDD2059 pKa = 3.23NFKK2062 pKa = 10.32IRR2064 pKa = 11.84HH2065 pKa = 6.24LDD2067 pKa = 3.04IQKK2070 pKa = 9.94PVAPTTVSLTNTATVTAPTGFTDD2093 pKa = 3.71TNTSNNSATDD2103 pKa = 3.44TDD2105 pKa = 4.48TVTAAPTVPSAPGVRR2120 pKa = 11.84TPGFWQNTEE2129 pKa = 3.64WQKK2132 pKa = 10.89FWDD2135 pKa = 5.33GIQGNEE2141 pKa = 4.28PAQKK2145 pKa = 8.27TQPNFAKK2152 pKa = 10.33SDD2154 pKa = 3.87LLFAPYY2160 pKa = 9.58TNSAQAGKK2168 pKa = 10.59VLDD2171 pKa = 4.33PVSGQYY2177 pKa = 10.67DD2178 pKa = 3.32IGLLIGDD2185 pKa = 4.45FNRR2188 pKa = 11.84NGKK2191 pKa = 7.69TDD2193 pKa = 3.35AGEE2196 pKa = 4.01DD2197 pKa = 3.46TLFYY2201 pKa = 10.77TRR2203 pKa = 11.84AQALQMVDD2211 pKa = 4.39ASAHH2215 pKa = 5.63PNGDD2219 pKa = 2.69KK2220 pKa = 10.61RR2221 pKa = 11.84YY2222 pKa = 9.23DD2223 pKa = 3.4LGRR2226 pKa = 11.84SLVAGWLNYY2235 pKa = 10.25LAGNPIDD2242 pKa = 3.98TANPTDD2248 pKa = 3.68KK2249 pKa = 11.14DD2250 pKa = 3.02EE2251 pKa = 4.64RR2252 pKa = 11.84YY2253 pKa = 10.42YY2254 pKa = 10.51INEE2257 pKa = 4.76GINWLQGFTPDD2268 pKa = 4.17EE2269 pKa = 4.51NGDD2272 pKa = 3.75KK2273 pKa = 10.95KK2274 pKa = 11.25GDD2276 pKa = 3.45GAIYY2280 pKa = 10.45QMTGSSVSSPQLTNSYY2296 pKa = 9.01WNLGISSASNLPSSYY2311 pKa = 10.82KK2312 pKa = 10.79SNTNVLYY2319 pKa = 9.51PLEE2322 pKa = 4.67SGSVINTNLDD2332 pKa = 3.28NYY2334 pKa = 11.31NNGLGLADD2342 pKa = 3.36NVFYY2346 pKa = 11.01GGNAA2350 pKa = 3.15
MM1 pKa = 7.51SRR3 pKa = 11.84PQVKK7 pKa = 8.45ITVSASGGPLNTPSYY22 pKa = 11.08KK23 pKa = 10.6DD24 pKa = 3.35LTITGIGTDD33 pKa = 3.27NTYY36 pKa = 9.32TFLNQQGVLDD46 pKa = 4.29AWCLDD51 pKa = 3.38SATGISSNTTYY62 pKa = 9.86TANVYY67 pKa = 10.52SSYY70 pKa = 11.25EE71 pKa = 3.96LGALATLGTPARR83 pKa = 11.84IGNLYY88 pKa = 10.75NLDD91 pKa = 3.66LVNWIINQNFAANPQYY107 pKa = 11.31NYY109 pKa = 11.33GEE111 pKa = 4.0IQAAIWTLLGDD122 pKa = 4.53TYY124 pKa = 11.37NSSGADD130 pKa = 3.34LTRR133 pKa = 11.84NGAVVASDD141 pKa = 3.64VNSIIALAKK150 pKa = 10.31ANGEE154 pKa = 4.04NFVPDD159 pKa = 3.71TNQFIGVIFDD169 pKa = 3.81PFVVNGDD176 pKa = 3.55GSVTPRR182 pKa = 11.84QPLIAQIKK190 pKa = 8.18AAKK193 pKa = 10.34LGDD196 pKa = 4.28LVWNDD201 pKa = 3.39LNANGIQDD209 pKa = 3.65TGEE212 pKa = 3.92SGIAGVTVNLTRR224 pKa = 11.84DD225 pKa = 3.52SNSNGIIEE233 pKa = 4.47SNEE236 pKa = 3.42ILATTTTNVSGLYY249 pKa = 9.12EE250 pKa = 4.26FRR252 pKa = 11.84GLTPGLSYY260 pKa = 10.35QVQFATPTGFDD271 pKa = 3.13AVSPRR276 pKa = 11.84QQGGNQTTDD285 pKa = 2.4SDD287 pKa = 4.27GLVSNVVILNPGEE300 pKa = 3.98FNQTIDD306 pKa = 3.17AGFYY310 pKa = 10.2KK311 pKa = 10.51YY312 pKa = 11.03ASLGDD317 pKa = 3.59FVFNDD322 pKa = 3.68GNNNGIQDD330 pKa = 3.76AGEE333 pKa = 4.05AGIGGVTVEE342 pKa = 5.58LINPANGSVIATTTTDD358 pKa = 3.82GIGAYY363 pKa = 8.44TFTGLTPGDD372 pKa = 3.82YY373 pKa = 9.53QVKK376 pKa = 8.04FTSPTGYY383 pKa = 10.33VFSTANVGDD392 pKa = 3.85DD393 pKa = 4.3TKK395 pKa = 11.6DD396 pKa = 3.11SDD398 pKa = 4.67ANTSTGLTKK407 pKa = 10.23TVTLTSGEE415 pKa = 4.31FNGTLDD421 pKa = 3.63AGLVQLASLGDD432 pKa = 3.93KK433 pKa = 10.5VFHH436 pKa = 7.25DD437 pKa = 5.01LDD439 pKa = 5.64ADD441 pKa = 5.05GIQDD445 pKa = 3.86AGEE448 pKa = 3.86QGIANASVKK457 pKa = 10.77LLDD460 pKa = 3.56TAGNVIATTTTDD472 pKa = 3.08GDD474 pKa = 3.85GLYY477 pKa = 10.64SFTNLQPGDD486 pKa = 3.73YY487 pKa = 9.59KK488 pKa = 11.37VQFVQPTGFNGVSLANVGGDD508 pKa = 4.01DD509 pKa = 4.78SVDD512 pKa = 3.57SDD514 pKa = 4.01GLISDD519 pKa = 4.49VVNLSPGEE527 pKa = 3.94NDD529 pKa = 3.38TTVDD533 pKa = 2.85AGFYY537 pKa = 8.57KK538 pKa = 10.21TASLGDD544 pKa = 3.5FVFNDD549 pKa = 3.68GNNNGIQDD557 pKa = 3.76AGEE560 pKa = 4.06AGIGEE565 pKa = 4.41VTVEE569 pKa = 5.02LINPADD575 pKa = 3.81GSVIATTTTDD585 pKa = 2.84EE586 pKa = 4.13TGAYY590 pKa = 7.47TFTGLTPGDD599 pKa = 3.82YY600 pKa = 9.53QVKK603 pKa = 8.25FTSPTDD609 pKa = 3.87YY610 pKa = 11.14VFSTADD616 pKa = 2.99QGGDD620 pKa = 3.44DD621 pKa = 4.5TVDD624 pKa = 3.2SDD626 pKa = 5.04ANTSTGLTQTVTLTSGEE643 pKa = 4.31FNGTLDD649 pKa = 3.63AGLVQLASLGDD660 pKa = 3.84FVFEE664 pKa = 4.86DD665 pKa = 3.94SNANGIQDD673 pKa = 3.09AWEE676 pKa = 3.81QGIAEE681 pKa = 4.27ASVNLLDD688 pKa = 3.78TAGNVIATTTTDD700 pKa = 3.08GDD702 pKa = 3.85GLYY705 pKa = 10.64SFTNLQPGDD714 pKa = 3.73YY715 pKa = 9.59KK716 pKa = 11.41VQFVQPAGFSGVSPSNVGGDD736 pKa = 3.97DD737 pKa = 4.02SVDD740 pKa = 3.57SDD742 pKa = 4.01GLISDD747 pKa = 4.49VVNLSPGEE755 pKa = 3.94NDD757 pKa = 3.38TTVDD761 pKa = 2.85AGFYY765 pKa = 8.57KK766 pKa = 10.21TASLGDD772 pKa = 3.5FVFNDD777 pKa = 3.68GNNNGIQDD785 pKa = 3.45IGEE788 pKa = 4.16AGIGGVTVEE797 pKa = 5.58LINPANGSVIASVITDD813 pKa = 3.89DD814 pKa = 3.48NGGYY818 pKa = 9.52QFSGLTPGDD827 pKa = 3.7YY828 pKa = 9.61QVKK831 pKa = 8.04FTSPTGYY838 pKa = 10.34VFSTAYY844 pKa = 10.15QGGDD848 pKa = 3.36DD849 pKa = 4.57TVDD852 pKa = 3.2SDD854 pKa = 5.04ANTSTGLTKK863 pKa = 10.23TVTLTSGEE871 pKa = 4.31FNGTLDD877 pKa = 3.63AGLVQLASLGDD888 pKa = 3.93KK889 pKa = 10.5VFHH892 pKa = 7.25DD893 pKa = 5.01LDD895 pKa = 5.64ADD897 pKa = 5.05GIQDD901 pKa = 3.86AGEE904 pKa = 3.86QGIANASVKK913 pKa = 10.77LLDD916 pKa = 3.56TAGNVIATTTTDD928 pKa = 3.08GDD930 pKa = 3.88GLYY933 pKa = 10.82SFINLQPGDD942 pKa = 3.83YY943 pKa = 9.82KK944 pKa = 11.37VQFVQPTDD952 pKa = 3.8FNGVSLANVGGDD964 pKa = 4.01DD965 pKa = 4.78SVDD968 pKa = 3.57SDD970 pKa = 4.01GLISDD975 pKa = 4.49VVNLSPGEE983 pKa = 3.94NDD985 pKa = 3.38TTVDD989 pKa = 2.85AGFYY993 pKa = 8.57KK994 pKa = 10.21TASLGDD1000 pKa = 3.5FVFNDD1005 pKa = 3.68GNNNGIQDD1013 pKa = 3.76AGEE1016 pKa = 4.05AGIGGVTVKK1025 pKa = 10.8LINPANGNVISTTTTDD1041 pKa = 3.03GAGAYY1046 pKa = 7.07TFTGLTPGDD1055 pKa = 3.84YY1056 pKa = 9.35QVKK1059 pKa = 9.7FISPTGYY1066 pKa = 10.67SFTAANQGGNDD1077 pKa = 4.28AVDD1080 pKa = 3.56SDD1082 pKa = 4.92ANPSTGLTQTVTLTSGEE1099 pKa = 4.32YY1100 pKa = 10.52NGTLDD1105 pKa = 4.27AGLVQLASLGDD1116 pKa = 3.93KK1117 pKa = 10.5VFHH1120 pKa = 7.25DD1121 pKa = 5.01LDD1123 pKa = 5.64ADD1125 pKa = 5.05GIQDD1129 pKa = 3.86AGEE1132 pKa = 3.86QGIANASVKK1141 pKa = 10.77LLDD1144 pKa = 3.56TAGNVIATTTTDD1156 pKa = 3.08GDD1158 pKa = 3.85GLYY1161 pKa = 10.64SFTNLQPGDD1170 pKa = 3.73YY1171 pKa = 9.59KK1172 pKa = 11.37VQFVQPTGFNGVSLANVGGDD1192 pKa = 3.78DD1193 pKa = 5.29SVDD1196 pKa = 3.18SDD1198 pKa = 3.89AASNLTTGVVNLSPGEE1214 pKa = 3.94NDD1216 pKa = 3.38TTVDD1220 pKa = 2.85AGFYY1224 pKa = 8.64KK1225 pKa = 10.25TASLGNFVFNDD1236 pKa = 3.34VNNNGIQEE1244 pKa = 3.98ASEE1247 pKa = 4.27AGVAGVTVTLTGGGADD1263 pKa = 3.84GLISTTADD1271 pKa = 2.81NTTITTTTNASGKK1284 pKa = 10.27YY1285 pKa = 9.23NFAGLTPGQEE1295 pKa = 4.26YY1296 pKa = 10.07QVGFSNLPAGYY1307 pKa = 10.36QFTQANAGTDD1317 pKa = 3.53DD1318 pKa = 5.35ALDD1321 pKa = 3.86SDD1323 pKa = 5.42ANTSTGKK1330 pKa = 7.77TQIVTLASGEE1340 pKa = 4.29NNTTLDD1346 pKa = 3.17AGIYY1350 pKa = 9.65QNAGDD1355 pKa = 4.69LSITKK1360 pKa = 9.53TNGLTTVSPGQQITYY1375 pKa = 9.2TIVAQNNGLITATNALVSDD1394 pKa = 5.22IIPSNLTNVTWTSVATGGATDD1415 pKa = 3.9NQSSGTGSINDD1426 pKa = 4.01YY1427 pKa = 9.5VTLTAGSSITYY1438 pKa = 8.23TVTGTVAPNALTPVYY1453 pKa = 8.18NTPTTINLTSNSSSALNGTAGNIRR1477 pKa = 11.84NFSANGISVNASAFSSTSTGTWNTAYY1503 pKa = 10.68LGLYY1507 pKa = 9.38GSYY1510 pKa = 11.06GLGVTDD1516 pKa = 4.27GGEE1519 pKa = 3.89NGSNGTHH1526 pKa = 7.02HH1527 pKa = 7.36IDD1529 pKa = 3.12NDD1531 pKa = 3.32GRR1533 pKa = 11.84NNYY1536 pKa = 9.11VLLEE1540 pKa = 4.14FSDD1543 pKa = 3.85SVVIDD1548 pKa = 3.95KK1549 pKa = 10.9AALSYY1554 pKa = 11.27VVGDD1558 pKa = 3.5SDD1560 pKa = 3.62ISVWIGNFNNPYY1572 pKa = 9.47TNHH1575 pKa = 6.17LTLNSSVLSSFGFTEE1590 pKa = 3.95INLGGSSDD1598 pKa = 3.79RR1599 pKa = 11.84NADD1602 pKa = 3.12INASNIVGNAVVIAAKK1618 pKa = 10.73VGDD1621 pKa = 3.98TNDD1624 pKa = 3.43EE1625 pKa = 4.37FKK1627 pKa = 11.13LKK1629 pKa = 10.81SLDD1632 pKa = 3.5IIKK1635 pKa = 9.23ATTQTGGSLVNTATITAPTGFTDD1658 pKa = 3.58TNSNNNSATDD1668 pKa = 3.23TDD1670 pKa = 4.38TIVASSTVRR1679 pKa = 11.84IGDD1682 pKa = 3.74RR1683 pKa = 11.84VWYY1686 pKa = 7.32DD1687 pKa = 3.32TNANGIQEE1695 pKa = 4.16TGEE1698 pKa = 4.0AGVAGVAVNLFNQAGTQVGTTTTDD1722 pKa = 2.83TNGFYY1727 pKa = 10.54QFTANTNSTYY1737 pKa = 10.71SVQFVQPTGFSGFTTANVGNDD1758 pKa = 3.61GADD1761 pKa = 3.24SDD1763 pKa = 4.55VVNANGTTAQFLVGTTDD1780 pKa = 3.26NLTIDD1785 pKa = 3.64AGLIKK1790 pKa = 10.76NIGDD1794 pKa = 3.81LSITKK1799 pKa = 9.48TDD1801 pKa = 3.16GLTTVTAGQQLTYY1814 pKa = 10.31TIVVSNNGFITATNALVTDD1833 pKa = 5.92AIPALLTNVTWTSVASGGATGNEE1856 pKa = 3.93TSGTGNINDD1865 pKa = 3.9YY1866 pKa = 9.57VTLTGGSSITYY1877 pKa = 7.49TVNGTVAANAVSSSSTYY1894 pKa = 9.84TDD1896 pKa = 2.52TRR1898 pKa = 11.84FDD1900 pKa = 4.14FNGNSSSDD1908 pKa = 3.44GTDD1911 pKa = 2.82GNTRR1915 pKa = 11.84TFTKK1919 pKa = 10.61DD1920 pKa = 2.88GITVTARR1927 pKa = 11.84AFSRR1931 pKa = 11.84VDD1933 pKa = 3.26GTNGAWSKK1941 pKa = 11.29AYY1943 pKa = 10.28LGSYY1947 pKa = 9.13TGGLGVTDD1955 pKa = 4.03SSEE1958 pKa = 3.83SSSYY1962 pKa = 10.79HH1963 pKa = 6.01RR1964 pKa = 11.84VDD1966 pKa = 3.43NGGGRR1971 pKa = 11.84DD1972 pKa = 3.58NYY1974 pKa = 10.9VLFQFSEE1981 pKa = 4.37AVVVDD1986 pKa = 3.89SAYY1989 pKa = 10.66LQYY1992 pKa = 11.4VVTDD1996 pKa = 3.36SDD1998 pKa = 3.82TSVWIGNFNNTITTLSDD2015 pKa = 3.22SVLNSFGFTEE2025 pKa = 4.4VNLGSGSDD2033 pKa = 3.11RR2034 pKa = 11.84WADD2037 pKa = 3.27INAGNYY2043 pKa = 9.46LGNTLVIAAKK2053 pKa = 9.9TDD2055 pKa = 3.43DD2056 pKa = 3.81TNDD2059 pKa = 3.23NFKK2062 pKa = 10.32IRR2064 pKa = 11.84HH2065 pKa = 6.24LDD2067 pKa = 3.04IQKK2070 pKa = 9.94PVAPTTVSLTNTATVTAPTGFTDD2093 pKa = 3.71TNTSNNSATDD2103 pKa = 3.44TDD2105 pKa = 4.48TVTAAPTVPSAPGVRR2120 pKa = 11.84TPGFWQNTEE2129 pKa = 3.64WQKK2132 pKa = 10.89FWDD2135 pKa = 5.33GIQGNEE2141 pKa = 4.28PAQKK2145 pKa = 8.27TQPNFAKK2152 pKa = 10.33SDD2154 pKa = 3.87LLFAPYY2160 pKa = 9.58TNSAQAGKK2168 pKa = 10.59VLDD2171 pKa = 4.33PVSGQYY2177 pKa = 10.67DD2178 pKa = 3.32IGLLIGDD2185 pKa = 4.45FNRR2188 pKa = 11.84NGKK2191 pKa = 7.69TDD2193 pKa = 3.35AGEE2196 pKa = 4.01DD2197 pKa = 3.46TLFYY2201 pKa = 10.77TRR2203 pKa = 11.84AQALQMVDD2211 pKa = 4.39ASAHH2215 pKa = 5.63PNGDD2219 pKa = 2.69KK2220 pKa = 10.61RR2221 pKa = 11.84YY2222 pKa = 9.23DD2223 pKa = 3.4LGRR2226 pKa = 11.84SLVAGWLNYY2235 pKa = 10.25LAGNPIDD2242 pKa = 3.98TANPTDD2248 pKa = 3.68KK2249 pKa = 11.14DD2250 pKa = 3.02EE2251 pKa = 4.64RR2252 pKa = 11.84YY2253 pKa = 10.42YY2254 pKa = 10.51INEE2257 pKa = 4.76GINWLQGFTPDD2268 pKa = 4.17EE2269 pKa = 4.51NGDD2272 pKa = 3.75KK2273 pKa = 10.95KK2274 pKa = 11.25GDD2276 pKa = 3.45GAIYY2280 pKa = 10.45QMTGSSVSSPQLTNSYY2296 pKa = 9.01WNLGISSASNLPSSYY2311 pKa = 10.82KK2312 pKa = 10.79SNTNVLYY2319 pKa = 9.51PLEE2322 pKa = 4.67SGSVINTNLDD2332 pKa = 3.28NYY2334 pKa = 11.31NNGLGLADD2342 pKa = 3.36NVFYY2346 pKa = 11.01GGNAA2350 pKa = 3.15
Molecular weight: 242.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A433UWE5|A0A433UWE5_ANAVA Uncharacterized protein OS=Trichormus variabilis SAG 1403-4b OX=447716 GN=DSM107003_12610 PE=4 SV=1
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84RR13 pKa = 11.84VGLKK17 pKa = 10.37EE18 pKa = 3.9EE19 pKa = 4.36FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 9.1VQLDD35 pKa = 3.24QGMGRR40 pKa = 11.84SAYY43 pKa = 9.32ICPQTSCLQAAQKK56 pKa = 10.5KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 5.08GTVPEE70 pKa = 4.21TLYY73 pKa = 9.23QTLWLRR79 pKa = 11.84LTQNNTQNQII89 pKa = 3.23
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84RR13 pKa = 11.84VGLKK17 pKa = 10.37EE18 pKa = 3.9EE19 pKa = 4.36FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 9.1VQLDD35 pKa = 3.24QGMGRR40 pKa = 11.84SAYY43 pKa = 9.32ICPQTSCLQAAQKK56 pKa = 10.5KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 5.08GTVPEE70 pKa = 4.21TLYY73 pKa = 9.23QTLWLRR79 pKa = 11.84LTQNNTQNQII89 pKa = 3.23
Molecular weight: 10.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1610029 |
41 |
3312 |
311.5 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.704 ± 0.033 | 0.971 ± 0.012 |
4.838 ± 0.027 | 6.374 ± 0.034 |
3.972 ± 0.023 | 6.499 ± 0.039 |
1.805 ± 0.016 | 7.239 ± 0.03 |
5.234 ± 0.034 | 10.926 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.834 ± 0.016 | 4.772 ± 0.036 |
4.565 ± 0.027 | 5.388 ± 0.034 |
4.73 ± 0.028 | 6.382 ± 0.03 |
5.753 ± 0.029 | 6.487 ± 0.028 |
1.408 ± 0.015 | 3.109 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
