
Shewanella maritima
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
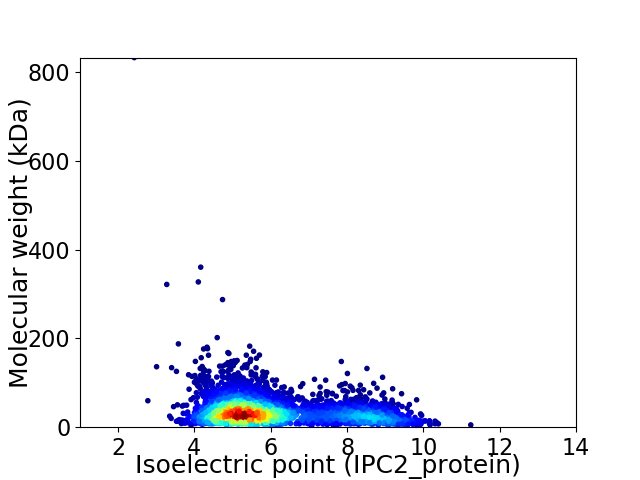
Virtual 2D-PAGE plot for 3709 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
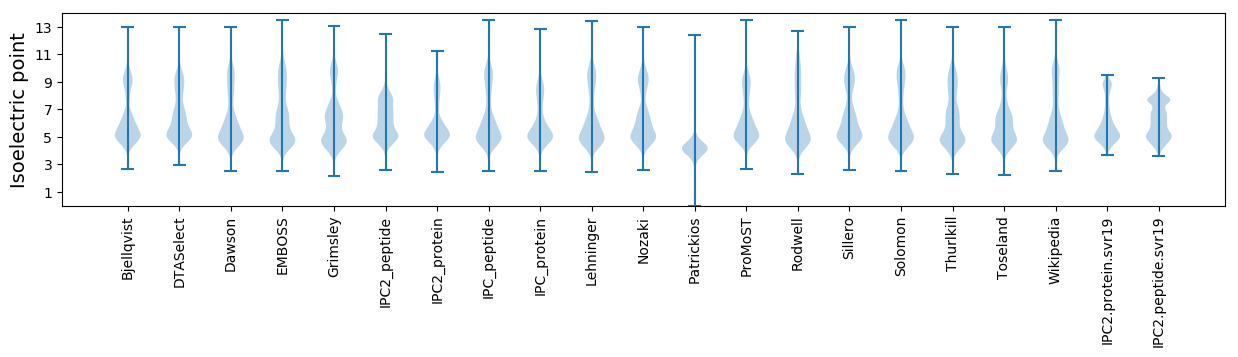
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A411PJ71|A0A411PJ71_9GAMM Response regulator OS=Shewanella maritima OX=2520507 GN=EXU30_13670 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.17RR3 pKa = 11.84IYY5 pKa = 10.05PIALAASLIALAGCSDD21 pKa = 4.59DD22 pKa = 5.28DD23 pKa = 4.45NDD25 pKa = 5.07VITIEE30 pKa = 3.9QDD32 pKa = 3.14EE33 pKa = 4.91PQTMAYY39 pKa = 9.88VRR41 pKa = 11.84VIHH44 pKa = 6.72AGSDD48 pKa = 3.47APKK51 pKa = 10.91VNVNVNGSQLLADD64 pKa = 3.83VDD66 pKa = 4.27YY67 pKa = 11.58AMSSGLIQVDD77 pKa = 3.51TATYY81 pKa = 10.26DD82 pKa = 3.21IDD84 pKa = 3.7VDD86 pKa = 4.95AILADD91 pKa = 3.72GTTATVLEE99 pKa = 4.73TNLSTEE105 pKa = 3.75ADD107 pKa = 3.44MEE109 pKa = 4.39YY110 pKa = 9.68TAVAVGSVAEE120 pKa = 4.12EE121 pKa = 3.82TLALKK126 pKa = 10.75LIANPMAAIASGYY139 pKa = 10.88ARR141 pKa = 11.84VQVLHH146 pKa = 6.61AAADD150 pKa = 3.59VGLVDD155 pKa = 5.05IYY157 pKa = 10.39VTEE160 pKa = 4.54PGADD164 pKa = 3.35IDD166 pKa = 4.16MMSPTLSANYY176 pKa = 8.94MDD178 pKa = 5.07NSEE181 pKa = 4.15QLEE184 pKa = 4.59LATGDD189 pKa = 3.47YY190 pKa = 10.16QIRR193 pKa = 11.84ITASGMKK200 pKa = 10.11DD201 pKa = 3.27VVYY204 pKa = 10.79DD205 pKa = 3.65SGTITLGDD213 pKa = 3.46MTDD216 pKa = 3.8YY217 pKa = 10.89LVSAITNTWSGDD229 pKa = 3.45APVALHH235 pKa = 6.27IALVEE240 pKa = 4.17GQVIVNDD247 pKa = 4.13ANSGADD253 pKa = 3.33LRR255 pKa = 11.84VIHH258 pKa = 6.69AVADD262 pKa = 3.87APEE265 pKa = 4.19VDD267 pKa = 4.23VFLDD271 pKa = 3.99DD272 pKa = 3.7ATSPAVDD279 pKa = 3.31MLAFKK284 pKa = 11.04AFTPYY289 pKa = 10.96VNIPEE294 pKa = 4.38GMHH297 pKa = 5.52TVTVAADD304 pKa = 3.5ADD306 pKa = 3.88NSVVVIDD313 pKa = 4.44KK314 pKa = 11.23APIEE318 pKa = 4.03PMIGMSYY325 pKa = 10.56SAFAVGSLSDD335 pKa = 5.19DD336 pKa = 3.63MIEE339 pKa = 4.14PLVLMEE345 pKa = 4.04DD346 pKa = 3.56TRR348 pKa = 11.84RR349 pKa = 11.84VATEE353 pKa = 3.39AKK355 pKa = 10.59LNVLHH360 pKa = 6.54SAYY363 pKa = 10.32SAPEE367 pKa = 3.49VDD369 pKa = 5.26IYY371 pKa = 11.01LTDD374 pKa = 3.55SADD377 pKa = 3.26ISEE380 pKa = 4.73ATPALTSVPFKK391 pKa = 9.91MASGSLSIAPGNYY404 pKa = 8.03TISVAVAGTKK414 pKa = 8.88TVAIGPLDD422 pKa = 3.59VTLEE426 pKa = 3.94AGGIYY431 pKa = 10.22GVAAVDD437 pKa = 3.66NMGGGAPFGVILLDD451 pKa = 4.36DD452 pKa = 5.23FNAEE456 pKa = 3.99MM457 pKa = 5.18
MM1 pKa = 7.34KK2 pKa = 10.17RR3 pKa = 11.84IYY5 pKa = 10.05PIALAASLIALAGCSDD21 pKa = 4.59DD22 pKa = 5.28DD23 pKa = 4.45NDD25 pKa = 5.07VITIEE30 pKa = 3.9QDD32 pKa = 3.14EE33 pKa = 4.91PQTMAYY39 pKa = 9.88VRR41 pKa = 11.84VIHH44 pKa = 6.72AGSDD48 pKa = 3.47APKK51 pKa = 10.91VNVNVNGSQLLADD64 pKa = 3.83VDD66 pKa = 4.27YY67 pKa = 11.58AMSSGLIQVDD77 pKa = 3.51TATYY81 pKa = 10.26DD82 pKa = 3.21IDD84 pKa = 3.7VDD86 pKa = 4.95AILADD91 pKa = 3.72GTTATVLEE99 pKa = 4.73TNLSTEE105 pKa = 3.75ADD107 pKa = 3.44MEE109 pKa = 4.39YY110 pKa = 9.68TAVAVGSVAEE120 pKa = 4.12EE121 pKa = 3.82TLALKK126 pKa = 10.75LIANPMAAIASGYY139 pKa = 10.88ARR141 pKa = 11.84VQVLHH146 pKa = 6.61AAADD150 pKa = 3.59VGLVDD155 pKa = 5.05IYY157 pKa = 10.39VTEE160 pKa = 4.54PGADD164 pKa = 3.35IDD166 pKa = 4.16MMSPTLSANYY176 pKa = 8.94MDD178 pKa = 5.07NSEE181 pKa = 4.15QLEE184 pKa = 4.59LATGDD189 pKa = 3.47YY190 pKa = 10.16QIRR193 pKa = 11.84ITASGMKK200 pKa = 10.11DD201 pKa = 3.27VVYY204 pKa = 10.79DD205 pKa = 3.65SGTITLGDD213 pKa = 3.46MTDD216 pKa = 3.8YY217 pKa = 10.89LVSAITNTWSGDD229 pKa = 3.45APVALHH235 pKa = 6.27IALVEE240 pKa = 4.17GQVIVNDD247 pKa = 4.13ANSGADD253 pKa = 3.33LRR255 pKa = 11.84VIHH258 pKa = 6.69AVADD262 pKa = 3.87APEE265 pKa = 4.19VDD267 pKa = 4.23VFLDD271 pKa = 3.99DD272 pKa = 3.7ATSPAVDD279 pKa = 3.31MLAFKK284 pKa = 11.04AFTPYY289 pKa = 10.96VNIPEE294 pKa = 4.38GMHH297 pKa = 5.52TVTVAADD304 pKa = 3.5ADD306 pKa = 3.88NSVVVIDD313 pKa = 4.44KK314 pKa = 11.23APIEE318 pKa = 4.03PMIGMSYY325 pKa = 10.56SAFAVGSLSDD335 pKa = 5.19DD336 pKa = 3.63MIEE339 pKa = 4.14PLVLMEE345 pKa = 4.04DD346 pKa = 3.56TRR348 pKa = 11.84RR349 pKa = 11.84VATEE353 pKa = 3.39AKK355 pKa = 10.59LNVLHH360 pKa = 6.54SAYY363 pKa = 10.32SAPEE367 pKa = 3.49VDD369 pKa = 5.26IYY371 pKa = 11.01LTDD374 pKa = 3.55SADD377 pKa = 3.26ISEE380 pKa = 4.73ATPALTSVPFKK391 pKa = 9.91MASGSLSIAPGNYY404 pKa = 8.03TISVAVAGTKK414 pKa = 8.88TVAIGPLDD422 pKa = 3.59VTLEE426 pKa = 3.94AGGIYY431 pKa = 10.22GVAAVDD437 pKa = 3.66NMGGGAPFGVILLDD451 pKa = 4.36DD452 pKa = 5.23FNAEE456 pKa = 3.99MM457 pKa = 5.18
Molecular weight: 47.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411PI14|A0A411PI14_9GAMM Der GTPase-activating protein YihI OS=Shewanella maritima OX=2520507 GN=yihI PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.01VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.01VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.39GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1257756 |
27 |
8132 |
339.1 |
37.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.186 ± 0.042 | 1.009 ± 0.016 |
5.797 ± 0.05 | 5.957 ± 0.043 |
3.994 ± 0.029 | 6.73 ± 0.038 |
2.228 ± 0.024 | 6.084 ± 0.031 |
5.256 ± 0.047 | 10.136 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.025 | 4.453 ± 0.048 |
3.784 ± 0.024 | 5.173 ± 0.044 |
4.124 ± 0.047 | 6.75 ± 0.04 |
5.47 ± 0.091 | 7.037 ± 0.037 |
1.171 ± 0.014 | 3.078 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
