
[Candida] pseudohaemulonii
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Metschnikowiaceae; Clavispora;
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
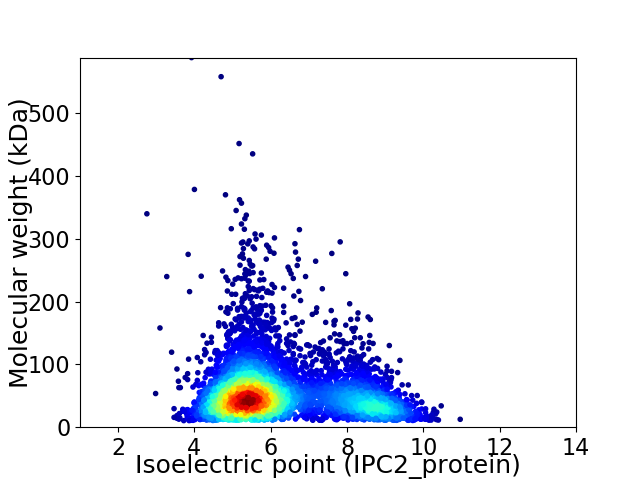
Virtual 2D-PAGE plot for 5132 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P7YNR6|A0A2P7YNR6_9ASCO Aminotran_1_2 domain-containing protein OS=[Candida] pseudohaemulonii OX=418784 GN=C7M61_003320 PE=3 SV=1
MM1 pKa = 7.43LAYY4 pKa = 10.19SVALFAYY11 pKa = 10.02AAVALGAQISGVFTSIDD28 pKa = 3.91SILAPANNRR37 pKa = 11.84PVTPSWQATVLWLMPRR53 pKa = 11.84LKK55 pKa = 10.51NIKK58 pKa = 10.43NGDD61 pKa = 3.65TFTLHH66 pKa = 5.54MPYY69 pKa = 10.31VFKK72 pKa = 10.02FTSLSNTLALTAGGTTFANCILFSGDD98 pKa = 3.14NVVGYY103 pKa = 10.18SEE105 pKa = 4.52VQCTATSAAEE115 pKa = 4.1NVEE118 pKa = 4.35SATGQITFPFTFNSGYY134 pKa = 10.82SLDD137 pKa = 4.01PVNLQAANAWNAGSNTVTWTDD158 pKa = 3.18GNTQISTTVNFLGGASQVILGSAEE182 pKa = 3.64NGVYY186 pKa = 10.77GLRR189 pKa = 11.84KK190 pKa = 8.86MVNLNVNQHH199 pKa = 5.57YY200 pKa = 10.72LLGPLCSSTMSGTLQITNPSSQVPFDD226 pKa = 4.06CLTIAGAISDD236 pKa = 4.89QINAWYY242 pKa = 10.0YY243 pKa = 8.09PQTANTVGVSVSSCSSTGAVVSFSNVPAGFRR274 pKa = 11.84PYY276 pKa = 10.24MNVNAAIPLGTFSSQNQYY294 pKa = 11.03SYY296 pKa = 11.57SFSCGWRR303 pKa = 11.84TLSGLITQLWTMYY316 pKa = 10.96SDD318 pKa = 4.05GNTGSGGAFKK328 pKa = 10.41PIVLTTVTDD337 pKa = 4.02PTATTTGVGTVTGASTNTIVVTVPISQTTITVTGTGTASTTHH379 pKa = 6.59SSTSSGTRR387 pKa = 11.84VISIDD392 pKa = 3.48VPTPTTTITRR402 pKa = 11.84TYY404 pKa = 10.32SGSTSSFLTVPATTGGTGTVIVDD427 pKa = 3.47VPTPTTTLTQTWTGTTTSTQTIPATSGGTATVIVDD462 pKa = 3.39VPTPVTTVTSTWTGLTTATVTQPVTSGGTQTVVVEE497 pKa = 4.57VPTPTTTLYY506 pKa = 9.46STWTGTVTTTTTVPATSGGTATVIVEE532 pKa = 4.27VPTPVTVITSTWTGSTTNVVTEE554 pKa = 4.16PFSLGTQTIVVEE566 pKa = 4.68VPTPTTTLTRR576 pKa = 11.84TWTGTEE582 pKa = 4.15TTTEE586 pKa = 4.35TIPAQSGGTGTVIVDD601 pKa = 3.27VPTPYY606 pKa = 8.22TTITRR611 pKa = 11.84TYY613 pKa = 8.32TGSVTTTEE621 pKa = 4.72TIPATTGGTGTVVVDD636 pKa = 3.64VPTPTTTITRR646 pKa = 11.84TWTGSTITTEE656 pKa = 4.25TVPATTGGTATLIVDD671 pKa = 4.23VPTPTTTVTSTYY683 pKa = 9.68TGLITTTVTVPATTGGTATVVVEE706 pKa = 4.52VPTPTTTITTTGDD719 pKa = 2.98VTNIVTLTILAASGGTEE736 pKa = 3.9TVVVEE741 pKa = 4.53VPTPTIYY748 pKa = 7.5TTRR751 pKa = 11.84TWTGSVTTTEE761 pKa = 4.72TIPATTGGTGTVIIDD776 pKa = 3.59VPTPTTTITRR786 pKa = 11.84TWTGSEE792 pKa = 4.17TTTEE796 pKa = 4.19TVPAASGGTGTVIVDD811 pKa = 3.48VPTPLTTITRR821 pKa = 11.84TWTGSITTTEE831 pKa = 4.4TIPAGSDD838 pKa = 3.11GTEE841 pKa = 4.09TIVVDD846 pKa = 4.03VPTPTTTITRR856 pKa = 11.84TWTGTEE862 pKa = 4.16STTEE866 pKa = 4.11TIPAQSGGTGTVIVDD881 pKa = 3.27VPTPYY886 pKa = 8.18TTITRR891 pKa = 11.84TWTGTTTQTEE901 pKa = 4.48TEE903 pKa = 4.16PAPSGGTGTVVIDD916 pKa = 3.7VPTPTTTITRR926 pKa = 11.84TWTGTEE932 pKa = 4.15TTTEE936 pKa = 4.36TIPAGSGGTEE946 pKa = 4.02TIIVDD951 pKa = 3.66VPTPYY956 pKa = 7.94TTLTRR961 pKa = 11.84TWTGTTTQTEE971 pKa = 4.48TEE973 pKa = 4.16PAPSGGTGTVVIDD986 pKa = 3.7VPTPTTTIYY995 pKa = 9.47STWTGSTISTRR1006 pKa = 11.84TIPASSGGTDD1016 pKa = 3.06TVVIEE1021 pKa = 4.6VPTPITVITSTWTGSITEE1039 pKa = 4.53VVTEE1043 pKa = 4.49PYY1045 pKa = 10.61EE1046 pKa = 4.65SGTQTIVVNVPTPTTTITRR1065 pKa = 11.84TWTGTEE1071 pKa = 4.11TTTEE1075 pKa = 4.15TVPAEE1080 pKa = 4.12SGGTGTVIIDD1090 pKa = 3.39VPTPYY1095 pKa = 7.93TTLTRR1100 pKa = 11.84TWTGTTTQTEE1110 pKa = 4.48TEE1112 pKa = 4.16PAPSGGTGTVIIDD1125 pKa = 3.59VPTPTTTITRR1135 pKa = 11.84TWTGTEE1141 pKa = 4.15TTTEE1145 pKa = 4.36TIPAGSGGTEE1155 pKa = 4.05TIVVDD1160 pKa = 3.87VPTPYY1165 pKa = 8.18TTITRR1170 pKa = 11.84TWTGTTTQTEE1180 pKa = 4.48TEE1182 pKa = 4.16PAPSGGTGTVVVDD1195 pKa = 3.48VPTPVTSITRR1205 pKa = 11.84TWTGTTTQTEE1215 pKa = 4.69TIPAQSGGTEE1225 pKa = 4.01TVIIDD1230 pKa = 3.58VPTPVTTLTRR1240 pKa = 11.84TWTDD1244 pKa = 3.1SITTTEE1250 pKa = 4.43TIPAEE1255 pKa = 4.22SGGTEE1260 pKa = 3.98TVIIDD1265 pKa = 3.73IPTPTTTLFSTWTGSTISTRR1285 pKa = 11.84TIPAEE1290 pKa = 4.02SGGTDD1295 pKa = 3.08TVVIEE1300 pKa = 4.6VPTPITVITSTWTGSITEE1318 pKa = 4.56IVTEE1322 pKa = 4.52PYY1324 pKa = 10.62EE1325 pKa = 4.7SGTQTIVVNVPTPVTTVTRR1344 pKa = 11.84TWTGTTTSTEE1354 pKa = 4.26TIPAEE1359 pKa = 4.17SGGTEE1364 pKa = 3.92TVIIDD1369 pKa = 3.58VPTPVTTITRR1379 pKa = 11.84TWTGTTTSTEE1389 pKa = 4.22TVPAEE1394 pKa = 4.1SGGTEE1399 pKa = 3.98TVIIDD1404 pKa = 3.44IPTPVTTITRR1414 pKa = 11.84TWTGTEE1420 pKa = 4.16STTEE1424 pKa = 4.12TIPAGSDD1431 pKa = 3.07GTEE1434 pKa = 3.9TVVVDD1439 pKa = 3.61VPTPYY1444 pKa = 8.15TTITRR1449 pKa = 11.84PWTGTFTTTEE1459 pKa = 4.15TEE1461 pKa = 4.07PAPSGGTGTVVVDD1474 pKa = 3.71VPYY1477 pKa = 10.71EE1478 pKa = 4.06ATTVTSTWTGTTTRR1492 pKa = 11.84TVSEE1496 pKa = 4.61PPAGSDD1502 pKa = 3.22TVGTVIIEE1510 pKa = 4.21VPPATTDD1517 pKa = 3.43YY1518 pKa = 7.55TTLTSTWTGTTTNTITNPPSGSDD1541 pKa = 3.2TVGTVVIEE1549 pKa = 4.12VPPTSSQFTTLTSTWTGTTTRR1570 pKa = 11.84TVTEE1574 pKa = 4.58PPSGSDD1580 pKa = 3.05TVGTVVIEE1588 pKa = 4.13VPPTSVDD1595 pKa = 3.44YY1596 pKa = 8.11TTVTSTWTGTTTATVTNPPSGSDD1619 pKa = 3.31TIGTVVIEE1627 pKa = 4.2VPPTSVDD1634 pKa = 3.44YY1635 pKa = 8.11TTVTSTWTGTTTATVTNPPSGSDD1658 pKa = 3.31TIGTVVIEE1666 pKa = 4.34VPPTSADD1673 pKa = 3.5YY1674 pKa = 7.52TTLTSTWTGTTTRR1687 pKa = 11.84TVTEE1691 pKa = 4.58PPSGSDD1697 pKa = 3.03TVGTVIIEE1705 pKa = 4.26VPPTTVPVIPPVLSWNWNSSYY1726 pKa = 10.87VVSLSSIPVPVSLSLSLTEE1745 pKa = 4.44SSEE1748 pKa = 4.15SSIASSIPPASGSEE1762 pKa = 3.92PATPAVSSSPASTEE1776 pKa = 3.88PPQLNTDD1783 pKa = 3.81SFSGPTSSGPITTASNQGPGPSPGPASDD1811 pKa = 3.87SSAPGPNPSGPNSGAEE1827 pKa = 4.27TSLSTASNPGPSPGPASEE1845 pKa = 4.33SSVPVSPGPNSEE1857 pKa = 4.31GSASTVTVVTTPGQPGPNPSGTGPVPSGPNSAGPSSSNPDD1897 pKa = 3.06TTVPVAPGPAPSGPAVPGTEE1917 pKa = 3.71SSAPGEE1923 pKa = 4.29SQPAPSGTANTEE1935 pKa = 4.14SQPSQTGPAPSGPNQTGPAPSGPNEE1960 pKa = 4.28TGSTPSGPNEE1970 pKa = 4.18NGPTSTGPAPANSAPPAPNPEE1991 pKa = 3.97PVASNTVPGSNPAPSGSNLGPAPSGPAPSGSNPGPVPVPGPAPSGTEE2038 pKa = 4.05SNNNPASTPYY2048 pKa = 7.94PTSVAGPSTRR2058 pKa = 11.84VPGNSAAASEE2068 pKa = 4.36TSSVVSEE2075 pKa = 4.81GPSSLVVCYY2084 pKa = 9.88KK2085 pKa = 10.29VAVSHH2090 pKa = 6.67ASSSSAPVGPEE2101 pKa = 3.74PSPASATPAISTYY2114 pKa = 10.85AGLAAAVKK2122 pKa = 10.06ISGLAAVVVPLLHH2135 pKa = 7.0VLFF2138 pKa = 5.26
MM1 pKa = 7.43LAYY4 pKa = 10.19SVALFAYY11 pKa = 10.02AAVALGAQISGVFTSIDD28 pKa = 3.91SILAPANNRR37 pKa = 11.84PVTPSWQATVLWLMPRR53 pKa = 11.84LKK55 pKa = 10.51NIKK58 pKa = 10.43NGDD61 pKa = 3.65TFTLHH66 pKa = 5.54MPYY69 pKa = 10.31VFKK72 pKa = 10.02FTSLSNTLALTAGGTTFANCILFSGDD98 pKa = 3.14NVVGYY103 pKa = 10.18SEE105 pKa = 4.52VQCTATSAAEE115 pKa = 4.1NVEE118 pKa = 4.35SATGQITFPFTFNSGYY134 pKa = 10.82SLDD137 pKa = 4.01PVNLQAANAWNAGSNTVTWTDD158 pKa = 3.18GNTQISTTVNFLGGASQVILGSAEE182 pKa = 3.64NGVYY186 pKa = 10.77GLRR189 pKa = 11.84KK190 pKa = 8.86MVNLNVNQHH199 pKa = 5.57YY200 pKa = 10.72LLGPLCSSTMSGTLQITNPSSQVPFDD226 pKa = 4.06CLTIAGAISDD236 pKa = 4.89QINAWYY242 pKa = 10.0YY243 pKa = 8.09PQTANTVGVSVSSCSSTGAVVSFSNVPAGFRR274 pKa = 11.84PYY276 pKa = 10.24MNVNAAIPLGTFSSQNQYY294 pKa = 11.03SYY296 pKa = 11.57SFSCGWRR303 pKa = 11.84TLSGLITQLWTMYY316 pKa = 10.96SDD318 pKa = 4.05GNTGSGGAFKK328 pKa = 10.41PIVLTTVTDD337 pKa = 4.02PTATTTGVGTVTGASTNTIVVTVPISQTTITVTGTGTASTTHH379 pKa = 6.59SSTSSGTRR387 pKa = 11.84VISIDD392 pKa = 3.48VPTPTTTITRR402 pKa = 11.84TYY404 pKa = 10.32SGSTSSFLTVPATTGGTGTVIVDD427 pKa = 3.47VPTPTTTLTQTWTGTTTSTQTIPATSGGTATVIVDD462 pKa = 3.39VPTPVTTVTSTWTGLTTATVTQPVTSGGTQTVVVEE497 pKa = 4.57VPTPTTTLYY506 pKa = 9.46STWTGTVTTTTTVPATSGGTATVIVEE532 pKa = 4.27VPTPVTVITSTWTGSTTNVVTEE554 pKa = 4.16PFSLGTQTIVVEE566 pKa = 4.68VPTPTTTLTRR576 pKa = 11.84TWTGTEE582 pKa = 4.15TTTEE586 pKa = 4.35TIPAQSGGTGTVIVDD601 pKa = 3.27VPTPYY606 pKa = 8.22TTITRR611 pKa = 11.84TYY613 pKa = 8.32TGSVTTTEE621 pKa = 4.72TIPATTGGTGTVVVDD636 pKa = 3.64VPTPTTTITRR646 pKa = 11.84TWTGSTITTEE656 pKa = 4.25TVPATTGGTATLIVDD671 pKa = 4.23VPTPTTTVTSTYY683 pKa = 9.68TGLITTTVTVPATTGGTATVVVEE706 pKa = 4.52VPTPTTTITTTGDD719 pKa = 2.98VTNIVTLTILAASGGTEE736 pKa = 3.9TVVVEE741 pKa = 4.53VPTPTIYY748 pKa = 7.5TTRR751 pKa = 11.84TWTGSVTTTEE761 pKa = 4.72TIPATTGGTGTVIIDD776 pKa = 3.59VPTPTTTITRR786 pKa = 11.84TWTGSEE792 pKa = 4.17TTTEE796 pKa = 4.19TVPAASGGTGTVIVDD811 pKa = 3.48VPTPLTTITRR821 pKa = 11.84TWTGSITTTEE831 pKa = 4.4TIPAGSDD838 pKa = 3.11GTEE841 pKa = 4.09TIVVDD846 pKa = 4.03VPTPTTTITRR856 pKa = 11.84TWTGTEE862 pKa = 4.16STTEE866 pKa = 4.11TIPAQSGGTGTVIVDD881 pKa = 3.27VPTPYY886 pKa = 8.18TTITRR891 pKa = 11.84TWTGTTTQTEE901 pKa = 4.48TEE903 pKa = 4.16PAPSGGTGTVVIDD916 pKa = 3.7VPTPTTTITRR926 pKa = 11.84TWTGTEE932 pKa = 4.15TTTEE936 pKa = 4.36TIPAGSGGTEE946 pKa = 4.02TIIVDD951 pKa = 3.66VPTPYY956 pKa = 7.94TTLTRR961 pKa = 11.84TWTGTTTQTEE971 pKa = 4.48TEE973 pKa = 4.16PAPSGGTGTVVIDD986 pKa = 3.7VPTPTTTIYY995 pKa = 9.47STWTGSTISTRR1006 pKa = 11.84TIPASSGGTDD1016 pKa = 3.06TVVIEE1021 pKa = 4.6VPTPITVITSTWTGSITEE1039 pKa = 4.53VVTEE1043 pKa = 4.49PYY1045 pKa = 10.61EE1046 pKa = 4.65SGTQTIVVNVPTPTTTITRR1065 pKa = 11.84TWTGTEE1071 pKa = 4.11TTTEE1075 pKa = 4.15TVPAEE1080 pKa = 4.12SGGTGTVIIDD1090 pKa = 3.39VPTPYY1095 pKa = 7.93TTLTRR1100 pKa = 11.84TWTGTTTQTEE1110 pKa = 4.48TEE1112 pKa = 4.16PAPSGGTGTVIIDD1125 pKa = 3.59VPTPTTTITRR1135 pKa = 11.84TWTGTEE1141 pKa = 4.15TTTEE1145 pKa = 4.36TIPAGSGGTEE1155 pKa = 4.05TIVVDD1160 pKa = 3.87VPTPYY1165 pKa = 8.18TTITRR1170 pKa = 11.84TWTGTTTQTEE1180 pKa = 4.48TEE1182 pKa = 4.16PAPSGGTGTVVVDD1195 pKa = 3.48VPTPVTSITRR1205 pKa = 11.84TWTGTTTQTEE1215 pKa = 4.69TIPAQSGGTEE1225 pKa = 4.01TVIIDD1230 pKa = 3.58VPTPVTTLTRR1240 pKa = 11.84TWTDD1244 pKa = 3.1SITTTEE1250 pKa = 4.43TIPAEE1255 pKa = 4.22SGGTEE1260 pKa = 3.98TVIIDD1265 pKa = 3.73IPTPTTTLFSTWTGSTISTRR1285 pKa = 11.84TIPAEE1290 pKa = 4.02SGGTDD1295 pKa = 3.08TVVIEE1300 pKa = 4.6VPTPITVITSTWTGSITEE1318 pKa = 4.56IVTEE1322 pKa = 4.52PYY1324 pKa = 10.62EE1325 pKa = 4.7SGTQTIVVNVPTPVTTVTRR1344 pKa = 11.84TWTGTTTSTEE1354 pKa = 4.26TIPAEE1359 pKa = 4.17SGGTEE1364 pKa = 3.92TVIIDD1369 pKa = 3.58VPTPVTTITRR1379 pKa = 11.84TWTGTTTSTEE1389 pKa = 4.22TVPAEE1394 pKa = 4.1SGGTEE1399 pKa = 3.98TVIIDD1404 pKa = 3.44IPTPVTTITRR1414 pKa = 11.84TWTGTEE1420 pKa = 4.16STTEE1424 pKa = 4.12TIPAGSDD1431 pKa = 3.07GTEE1434 pKa = 3.9TVVVDD1439 pKa = 3.61VPTPYY1444 pKa = 8.15TTITRR1449 pKa = 11.84PWTGTFTTTEE1459 pKa = 4.15TEE1461 pKa = 4.07PAPSGGTGTVVVDD1474 pKa = 3.71VPYY1477 pKa = 10.71EE1478 pKa = 4.06ATTVTSTWTGTTTRR1492 pKa = 11.84TVSEE1496 pKa = 4.61PPAGSDD1502 pKa = 3.22TVGTVIIEE1510 pKa = 4.21VPPATTDD1517 pKa = 3.43YY1518 pKa = 7.55TTLTSTWTGTTTNTITNPPSGSDD1541 pKa = 3.2TVGTVVIEE1549 pKa = 4.12VPPTSSQFTTLTSTWTGTTTRR1570 pKa = 11.84TVTEE1574 pKa = 4.58PPSGSDD1580 pKa = 3.05TVGTVVIEE1588 pKa = 4.13VPPTSVDD1595 pKa = 3.44YY1596 pKa = 8.11TTVTSTWTGTTTATVTNPPSGSDD1619 pKa = 3.31TIGTVVIEE1627 pKa = 4.2VPPTSVDD1634 pKa = 3.44YY1635 pKa = 8.11TTVTSTWTGTTTATVTNPPSGSDD1658 pKa = 3.31TIGTVVIEE1666 pKa = 4.34VPPTSADD1673 pKa = 3.5YY1674 pKa = 7.52TTLTSTWTGTTTRR1687 pKa = 11.84TVTEE1691 pKa = 4.58PPSGSDD1697 pKa = 3.03TVGTVIIEE1705 pKa = 4.26VPPTTVPVIPPVLSWNWNSSYY1726 pKa = 10.87VVSLSSIPVPVSLSLSLTEE1745 pKa = 4.44SSEE1748 pKa = 4.15SSIASSIPPASGSEE1762 pKa = 3.92PATPAVSSSPASTEE1776 pKa = 3.88PPQLNTDD1783 pKa = 3.81SFSGPTSSGPITTASNQGPGPSPGPASDD1811 pKa = 3.87SSAPGPNPSGPNSGAEE1827 pKa = 4.27TSLSTASNPGPSPGPASEE1845 pKa = 4.33SSVPVSPGPNSEE1857 pKa = 4.31GSASTVTVVTTPGQPGPNPSGTGPVPSGPNSAGPSSSNPDD1897 pKa = 3.06TTVPVAPGPAPSGPAVPGTEE1917 pKa = 3.71SSAPGEE1923 pKa = 4.29SQPAPSGTANTEE1935 pKa = 4.14SQPSQTGPAPSGPNQTGPAPSGPNEE1960 pKa = 4.28TGSTPSGPNEE1970 pKa = 4.18NGPTSTGPAPANSAPPAPNPEE1991 pKa = 3.97PVASNTVPGSNPAPSGSNLGPAPSGPAPSGSNPGPVPVPGPAPSGTEE2038 pKa = 4.05SNNNPASTPYY2048 pKa = 7.94PTSVAGPSTRR2058 pKa = 11.84VPGNSAAASEE2068 pKa = 4.36TSSVVSEE2075 pKa = 4.81GPSSLVVCYY2084 pKa = 9.88KK2085 pKa = 10.29VAVSHH2090 pKa = 6.67ASSSSAPVGPEE2101 pKa = 3.74PSPASATPAISTYY2114 pKa = 10.85AGLAAAVKK2122 pKa = 10.06ISGLAAVVVPLLHH2135 pKa = 7.0VLFF2138 pKa = 5.26
Molecular weight: 215.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P7YRU4|A0A2P7YRU4_9ASCO Uncharacterized protein OS=[Candida] pseudohaemulonii OX=418784 GN=C7M61_002605 PE=4 SV=1
MM1 pKa = 7.37FAGLKK6 pKa = 9.73HH7 pKa = 6.02CQNSLRR13 pKa = 11.84VGLNATRR20 pKa = 11.84QFSMLNKK27 pKa = 9.86PLITTSLSSSPISRR41 pKa = 11.84MPVQIPQPSTLSLLLNLIQRR61 pKa = 11.84RR62 pKa = 11.84FKK64 pKa = 11.22SRR66 pKa = 11.84GNTYY70 pKa = 10.24QPSTLKK76 pKa = 10.47RR77 pKa = 11.84KK78 pKa = 8.63RR79 pKa = 11.84TFGFLARR86 pKa = 11.84LRR88 pKa = 11.84SRR90 pKa = 11.84GGRR93 pKa = 11.84KK94 pKa = 8.67ILQRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 10.5AKK102 pKa = 9.78GRR104 pKa = 11.84WYY106 pKa = 9.08LTHH109 pKa = 7.23
MM1 pKa = 7.37FAGLKK6 pKa = 9.73HH7 pKa = 6.02CQNSLRR13 pKa = 11.84VGLNATRR20 pKa = 11.84QFSMLNKK27 pKa = 9.86PLITTSLSSSPISRR41 pKa = 11.84MPVQIPQPSTLSLLLNLIQRR61 pKa = 11.84RR62 pKa = 11.84FKK64 pKa = 11.22SRR66 pKa = 11.84GNTYY70 pKa = 10.24QPSTLKK76 pKa = 10.47RR77 pKa = 11.84KK78 pKa = 8.63RR79 pKa = 11.84TFGFLARR86 pKa = 11.84LRR88 pKa = 11.84SRR90 pKa = 11.84GGRR93 pKa = 11.84KK94 pKa = 8.67ILQRR98 pKa = 11.84RR99 pKa = 11.84KK100 pKa = 10.5AKK102 pKa = 9.78GRR104 pKa = 11.84WYY106 pKa = 9.08LTHH109 pKa = 7.23
Molecular weight: 12.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2713752 |
99 |
5748 |
528.8 |
59.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.813 ± 0.032 | 1.084 ± 0.011 |
6.012 ± 0.026 | 6.985 ± 0.035 |
4.43 ± 0.027 | 5.584 ± 0.036 |
2.23 ± 0.014 | 5.444 ± 0.025 |
6.807 ± 0.035 | 10.234 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.013 | 4.765 ± 0.022 |
4.87 ± 0.03 | 3.976 ± 0.025 |
4.621 ± 0.026 | 7.978 ± 0.033 |
5.539 ± 0.07 | 6.317 ± 0.028 |
1.097 ± 0.011 | 3.227 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
