
Nostoc minutum NIES-26
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; Nostoc minutum
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
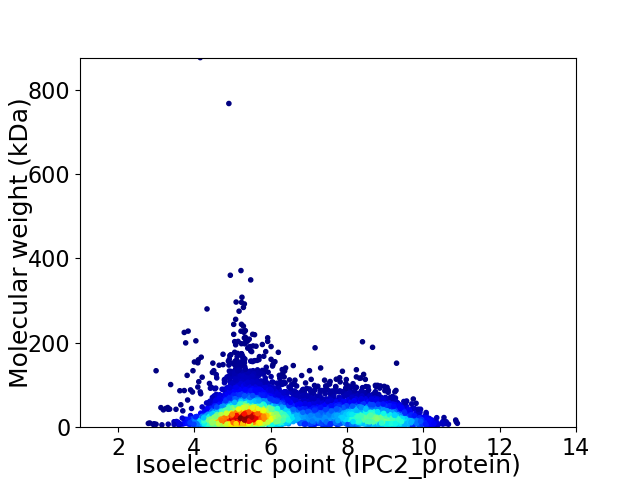
Virtual 2D-PAGE plot for 7588 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367QJY6|A0A367QJY6_9NOSO HTH tetR-type domain-containing protein OS=Nostoc minutum NIES-26 OX=1844469 GN=A6770_28270 PE=4 SV=1
MM1 pKa = 7.45PVFVGTLFNDD11 pKa = 3.59TLTGYY16 pKa = 10.82VYY18 pKa = 11.16DD19 pKa = 4.47DD20 pKa = 4.17TLSGLAGNDD29 pKa = 2.92ILLGKK34 pKa = 10.0AGNDD38 pKa = 3.88DD39 pKa = 3.25ISGGEE44 pKa = 4.01GSDD47 pKa = 3.58WLDD50 pKa = 2.91GDD52 pKa = 4.67AGNDD56 pKa = 3.67TLRR59 pKa = 11.84GDD61 pKa = 3.74PFISEE66 pKa = 4.76DD67 pKa = 3.41EE68 pKa = 4.29DD69 pKa = 4.14EE70 pKa = 4.66ILRR73 pKa = 11.84LSPKK77 pKa = 10.47ARR79 pKa = 11.84DD80 pKa = 3.57TLRR83 pKa = 11.84GGLGNDD89 pKa = 3.74LLEE92 pKa = 4.97GGAGKK97 pKa = 8.63DD98 pKa = 3.48TLIGSAGDD106 pKa = 3.54DD107 pKa = 4.05VLWGGYY113 pKa = 8.8NDD115 pKa = 4.01YY116 pKa = 11.35EE117 pKa = 4.44GVLPFSSGDD126 pKa = 3.27YY127 pKa = 10.74GSFIEE132 pKa = 5.48KK133 pKa = 10.92YY134 pKa = 10.05NDD136 pKa = 3.26NGNDD140 pKa = 3.79LLVGGSGNDD149 pKa = 3.56TLHH152 pKa = 6.91GDD154 pKa = 4.35SIFADD159 pKa = 3.12ILFNGVFAIGGNDD172 pKa = 3.47TLDD175 pKa = 3.79GGSGNDD181 pKa = 3.81LLEE184 pKa = 4.67GSLGKK189 pKa = 10.35DD190 pKa = 3.23YY191 pKa = 11.2LYY193 pKa = 11.12GGSGNDD199 pKa = 3.37RR200 pKa = 11.84IYY202 pKa = 11.27GGTLNPNRR210 pKa = 11.84RR211 pKa = 11.84QSDD214 pKa = 3.25AGDD217 pKa = 3.3DD218 pKa = 3.68HH219 pKa = 8.48LYY221 pKa = 10.93GGSGDD226 pKa = 4.56DD227 pKa = 4.66LLAASIVDD235 pKa = 3.53IYY237 pKa = 10.9FGSGRR242 pKa = 11.84NDD244 pKa = 3.33FLDD247 pKa = 4.17GGAGNDD253 pKa = 3.65TLYY256 pKa = 11.46GEE258 pKa = 4.9AGSDD262 pKa = 3.76FLNGGAGNDD271 pKa = 3.66VLIGDD276 pKa = 3.78TDD278 pKa = 4.1FVFFRR283 pKa = 11.84QNTVDD288 pKa = 4.13VVDD291 pKa = 4.3TLTGGAGSDD300 pKa = 3.6QFILGSDD307 pKa = 3.46VHH309 pKa = 7.17GLTLYY314 pKa = 10.9GLAFGEE320 pKa = 4.16EE321 pKa = 4.56STFTEE326 pKa = 4.31YY327 pKa = 11.0AIITDD332 pKa = 4.63FNTNQDD338 pKa = 3.75IIQLAKK344 pKa = 10.45LYY346 pKa = 10.17YY347 pKa = 9.85PSDD350 pKa = 3.47LGLYY354 pKa = 9.46YY355 pKa = 9.9PANYY359 pKa = 9.97VLGSAPVGQTGTGIYY374 pKa = 9.35IDD376 pKa = 3.89QEE378 pKa = 4.01EE379 pKa = 4.25QADD382 pKa = 3.82VLVAVVQGVSNLSLSEE398 pKa = 4.17SYY400 pKa = 10.84FRR402 pKa = 11.84FVV404 pKa = 3.43
MM1 pKa = 7.45PVFVGTLFNDD11 pKa = 3.59TLTGYY16 pKa = 10.82VYY18 pKa = 11.16DD19 pKa = 4.47DD20 pKa = 4.17TLSGLAGNDD29 pKa = 2.92ILLGKK34 pKa = 10.0AGNDD38 pKa = 3.88DD39 pKa = 3.25ISGGEE44 pKa = 4.01GSDD47 pKa = 3.58WLDD50 pKa = 2.91GDD52 pKa = 4.67AGNDD56 pKa = 3.67TLRR59 pKa = 11.84GDD61 pKa = 3.74PFISEE66 pKa = 4.76DD67 pKa = 3.41EE68 pKa = 4.29DD69 pKa = 4.14EE70 pKa = 4.66ILRR73 pKa = 11.84LSPKK77 pKa = 10.47ARR79 pKa = 11.84DD80 pKa = 3.57TLRR83 pKa = 11.84GGLGNDD89 pKa = 3.74LLEE92 pKa = 4.97GGAGKK97 pKa = 8.63DD98 pKa = 3.48TLIGSAGDD106 pKa = 3.54DD107 pKa = 4.05VLWGGYY113 pKa = 8.8NDD115 pKa = 4.01YY116 pKa = 11.35EE117 pKa = 4.44GVLPFSSGDD126 pKa = 3.27YY127 pKa = 10.74GSFIEE132 pKa = 5.48KK133 pKa = 10.92YY134 pKa = 10.05NDD136 pKa = 3.26NGNDD140 pKa = 3.79LLVGGSGNDD149 pKa = 3.56TLHH152 pKa = 6.91GDD154 pKa = 4.35SIFADD159 pKa = 3.12ILFNGVFAIGGNDD172 pKa = 3.47TLDD175 pKa = 3.79GGSGNDD181 pKa = 3.81LLEE184 pKa = 4.67GSLGKK189 pKa = 10.35DD190 pKa = 3.23YY191 pKa = 11.2LYY193 pKa = 11.12GGSGNDD199 pKa = 3.37RR200 pKa = 11.84IYY202 pKa = 11.27GGTLNPNRR210 pKa = 11.84RR211 pKa = 11.84QSDD214 pKa = 3.25AGDD217 pKa = 3.3DD218 pKa = 3.68HH219 pKa = 8.48LYY221 pKa = 10.93GGSGDD226 pKa = 4.56DD227 pKa = 4.66LLAASIVDD235 pKa = 3.53IYY237 pKa = 10.9FGSGRR242 pKa = 11.84NDD244 pKa = 3.33FLDD247 pKa = 4.17GGAGNDD253 pKa = 3.65TLYY256 pKa = 11.46GEE258 pKa = 4.9AGSDD262 pKa = 3.76FLNGGAGNDD271 pKa = 3.66VLIGDD276 pKa = 3.78TDD278 pKa = 4.1FVFFRR283 pKa = 11.84QNTVDD288 pKa = 4.13VVDD291 pKa = 4.3TLTGGAGSDD300 pKa = 3.6QFILGSDD307 pKa = 3.46VHH309 pKa = 7.17GLTLYY314 pKa = 10.9GLAFGEE320 pKa = 4.16EE321 pKa = 4.56STFTEE326 pKa = 4.31YY327 pKa = 11.0AIITDD332 pKa = 4.63FNTNQDD338 pKa = 3.75IIQLAKK344 pKa = 10.45LYY346 pKa = 10.17YY347 pKa = 9.85PSDD350 pKa = 3.47LGLYY354 pKa = 9.46YY355 pKa = 9.9PANYY359 pKa = 9.97VLGSAPVGQTGTGIYY374 pKa = 9.35IDD376 pKa = 3.89QEE378 pKa = 4.01EE379 pKa = 4.25QADD382 pKa = 3.82VLVAVVQGVSNLSLSEE398 pKa = 4.17SYY400 pKa = 10.84FRR402 pKa = 11.84FVV404 pKa = 3.43
Molecular weight: 42.38 kDa
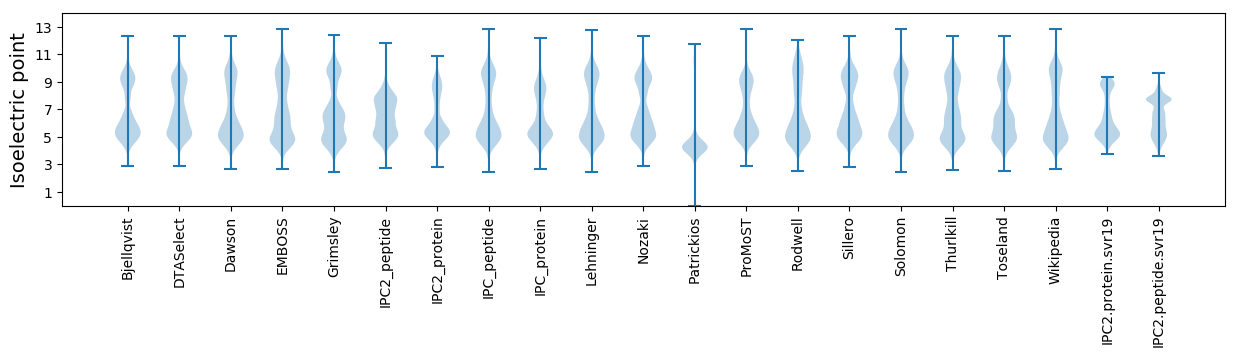
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A367S1R7|A0A367S1R7_9NOSO Acetyltransferase OS=Nostoc minutum NIES-26 OX=1844469 GN=A6770_35930 PE=4 SV=1
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84KK13 pKa = 9.35VSSKK17 pKa = 10.83EE18 pKa = 3.79DD19 pKa = 3.04FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 8.66VQLDD35 pKa = 3.07RR36 pKa = 11.84GMGRR40 pKa = 11.84SAYY43 pKa = 9.33ICPQPGCLQAAQKK56 pKa = 10.49KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 6.06ASVPEE70 pKa = 4.0TLYY73 pKa = 9.09QTLWQRR79 pKa = 11.84LAA81 pKa = 3.68
MM1 pKa = 7.31KK2 pKa = 10.03PNYY5 pKa = 9.07RR6 pKa = 11.84RR7 pKa = 11.84CISCRR12 pKa = 11.84KK13 pKa = 9.35VSSKK17 pKa = 10.83EE18 pKa = 3.79DD19 pKa = 3.04FWRR22 pKa = 11.84IVRR25 pKa = 11.84VFPSGKK31 pKa = 8.66VQLDD35 pKa = 3.07RR36 pKa = 11.84GMGRR40 pKa = 11.84SAYY43 pKa = 9.33ICPQPGCLQAAQKK56 pKa = 10.49KK57 pKa = 8.83NRR59 pKa = 11.84LGRR62 pKa = 11.84SLHH65 pKa = 6.06ASVPEE70 pKa = 4.0TLYY73 pKa = 9.09QTLWQRR79 pKa = 11.84LAA81 pKa = 3.68
Molecular weight: 9.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2454932 |
31 |
8449 |
323.5 |
36.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.123 ± 0.03 | 0.996 ± 0.01 |
4.875 ± 0.024 | 6.275 ± 0.03 |
3.904 ± 0.018 | 6.49 ± 0.032 |
1.821 ± 0.014 | 6.735 ± 0.025 |
4.97 ± 0.028 | 10.965 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.721 ± 0.013 | 4.5 ± 0.026 |
4.574 ± 0.023 | 5.558 ± 0.027 |
5.112 ± 0.023 | 6.508 ± 0.026 |
5.745 ± 0.027 | 6.62 ± 0.018 |
1.43 ± 0.012 | 3.075 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
