
Lawsonibacter asaccharolyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Lawsonibacter
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
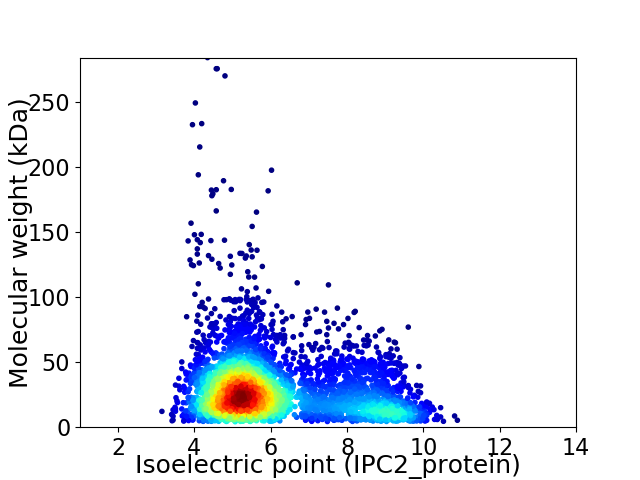
Virtual 2D-PAGE plot for 4274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P2FE84|A0A2P2FE84_9FIRM Metallophos domain-containing protein OS=Lawsonibacter asaccharolyticus OX=2108523 GN=LAWASA_4327 PE=4 SV=1
MM1 pKa = 7.35EE2 pKa = 6.15HH3 pKa = 7.55DD4 pKa = 3.71SMDD7 pKa = 3.43RR8 pKa = 11.84KK9 pKa = 10.68LSVLLCGALTLALAGCQAPVEE30 pKa = 4.41EE31 pKa = 5.0PLLDD35 pKa = 3.4PTVTVEE41 pKa = 4.29TTAVQAGDD49 pKa = 4.36LSTQSTYY56 pKa = 10.83IGTISAEE63 pKa = 4.09GTASVVSMISGNVEE77 pKa = 3.76QVAVAVGDD85 pKa = 3.96TVSAGDD91 pKa = 3.8LLCRR95 pKa = 11.84FDD97 pKa = 5.63DD98 pKa = 4.21EE99 pKa = 4.68SARR102 pKa = 11.84LSLQNAQAAANSAQEE117 pKa = 4.55SYY119 pKa = 11.44NSAVSNYY126 pKa = 10.13GGTDD130 pKa = 3.38LPLLQEE136 pKa = 4.3QLRR139 pKa = 11.84LAQDD143 pKa = 3.6NYY145 pKa = 11.61DD146 pKa = 3.61DD147 pKa = 4.01TLALLEE153 pKa = 4.78IGAASQVEE161 pKa = 4.42VDD163 pKa = 3.81QAHH166 pKa = 5.0QTLLSAQAGLEE177 pKa = 3.93AAQASLNSARR187 pKa = 11.84AGIQSAQVGVDD198 pKa = 3.15SAQYY202 pKa = 10.55QLSLYY207 pKa = 10.7NLTAPINGVVEE218 pKa = 4.45AVNVTQNNFAASGQVAFVISNGNNKK243 pKa = 8.14TVTFYY248 pKa = 10.48VTDD251 pKa = 3.64SVRR254 pKa = 11.84QTLTPGQAVTVTYY267 pKa = 10.65NGQSYY272 pKa = 9.94EE273 pKa = 4.18GAVTEE278 pKa = 4.07IGGVVDD284 pKa = 3.84AQVGQFRR291 pKa = 11.84VKK293 pKa = 10.76AIIDD297 pKa = 3.88GAQDD301 pKa = 3.83LPDD304 pKa = 3.95GLAVEE309 pKa = 5.07LTTTAHH315 pKa = 5.26QVTGAVLIPSDD326 pKa = 3.69AMFFEE331 pKa = 5.04SGVSYY336 pKa = 10.72VYY338 pKa = 10.64VVQDD342 pKa = 3.49GKK344 pKa = 10.92AVRR347 pKa = 11.84SDD349 pKa = 3.5VTIGLYY355 pKa = 9.36TADD358 pKa = 4.39TIAVTSGLTLGDD370 pKa = 3.71EE371 pKa = 5.32VITTWSSSLRR381 pKa = 11.84DD382 pKa = 3.55GVSVQTAAQTEE393 pKa = 4.35DD394 pKa = 3.87SEE396 pKa = 5.03SAQADD401 pKa = 3.74PAASGPEE408 pKa = 3.88AEE410 pKa = 4.66GG411 pKa = 3.31
MM1 pKa = 7.35EE2 pKa = 6.15HH3 pKa = 7.55DD4 pKa = 3.71SMDD7 pKa = 3.43RR8 pKa = 11.84KK9 pKa = 10.68LSVLLCGALTLALAGCQAPVEE30 pKa = 4.41EE31 pKa = 5.0PLLDD35 pKa = 3.4PTVTVEE41 pKa = 4.29TTAVQAGDD49 pKa = 4.36LSTQSTYY56 pKa = 10.83IGTISAEE63 pKa = 4.09GTASVVSMISGNVEE77 pKa = 3.76QVAVAVGDD85 pKa = 3.96TVSAGDD91 pKa = 3.8LLCRR95 pKa = 11.84FDD97 pKa = 5.63DD98 pKa = 4.21EE99 pKa = 4.68SARR102 pKa = 11.84LSLQNAQAAANSAQEE117 pKa = 4.55SYY119 pKa = 11.44NSAVSNYY126 pKa = 10.13GGTDD130 pKa = 3.38LPLLQEE136 pKa = 4.3QLRR139 pKa = 11.84LAQDD143 pKa = 3.6NYY145 pKa = 11.61DD146 pKa = 3.61DD147 pKa = 4.01TLALLEE153 pKa = 4.78IGAASQVEE161 pKa = 4.42VDD163 pKa = 3.81QAHH166 pKa = 5.0QTLLSAQAGLEE177 pKa = 3.93AAQASLNSARR187 pKa = 11.84AGIQSAQVGVDD198 pKa = 3.15SAQYY202 pKa = 10.55QLSLYY207 pKa = 10.7NLTAPINGVVEE218 pKa = 4.45AVNVTQNNFAASGQVAFVISNGNNKK243 pKa = 8.14TVTFYY248 pKa = 10.48VTDD251 pKa = 3.64SVRR254 pKa = 11.84QTLTPGQAVTVTYY267 pKa = 10.65NGQSYY272 pKa = 9.94EE273 pKa = 4.18GAVTEE278 pKa = 4.07IGGVVDD284 pKa = 3.84AQVGQFRR291 pKa = 11.84VKK293 pKa = 10.76AIIDD297 pKa = 3.88GAQDD301 pKa = 3.83LPDD304 pKa = 3.95GLAVEE309 pKa = 5.07LTTTAHH315 pKa = 5.26QVTGAVLIPSDD326 pKa = 3.69AMFFEE331 pKa = 5.04SGVSYY336 pKa = 10.72VYY338 pKa = 10.64VVQDD342 pKa = 3.49GKK344 pKa = 10.92AVRR347 pKa = 11.84SDD349 pKa = 3.5VTIGLYY355 pKa = 9.36TADD358 pKa = 4.39TIAVTSGLTLGDD370 pKa = 3.71EE371 pKa = 5.32VITTWSSSLRR381 pKa = 11.84DD382 pKa = 3.55GVSVQTAAQTEE393 pKa = 4.35DD394 pKa = 3.87SEE396 pKa = 5.03SAQADD401 pKa = 3.74PAASGPEE408 pKa = 3.88AEE410 pKa = 4.66GG411 pKa = 3.31
Molecular weight: 42.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P2F7H2|A0A2P2F7H2_9FIRM Uncharacterized protein OS=Lawsonibacter asaccharolyticus OX=2108523 GN=LAWASA_1982 PE=4 SV=1
MM1 pKa = 7.23TGSLAGAWGVAVMVALAALLLLHH24 pKa = 6.88RR25 pKa = 11.84PLGRR29 pKa = 11.84LLRR32 pKa = 11.84LAVRR36 pKa = 11.84SSMGLAALALLNQIGLGLGVNLVNALILGVLGVPGLGLLLMLQWVLRR83 pKa = 11.84TT84 pKa = 3.72
MM1 pKa = 7.23TGSLAGAWGVAVMVALAALLLLHH24 pKa = 6.88RR25 pKa = 11.84PLGRR29 pKa = 11.84LLRR32 pKa = 11.84LAVRR36 pKa = 11.84SSMGLAALALLNQIGLGLGVNLVNALILGVLGVPGLGLLLMLQWVLRR83 pKa = 11.84TT84 pKa = 3.72
Molecular weight: 8.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1174145 |
39 |
2626 |
274.7 |
30.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.295 ± 0.046 | 1.593 ± 0.019 |
5.641 ± 0.037 | 7.036 ± 0.044 |
3.676 ± 0.028 | 8.009 ± 0.036 |
1.788 ± 0.021 | 5.346 ± 0.033 |
4.447 ± 0.042 | 10.114 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.809 ± 0.021 | 3.096 ± 0.03 |
4.4 ± 0.029 | 3.77 ± 0.025 |
6.064 ± 0.044 | 5.736 ± 0.032 |
5.449 ± 0.049 | 7.027 ± 0.037 |
1.21 ± 0.017 | 3.494 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
