
Blumeria graminis f. sp. tritici 96224
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Erysiphales; Erysiphaceae; Blumeria; Blumeria graminis; Blumeria graminis f. sp. tritici
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
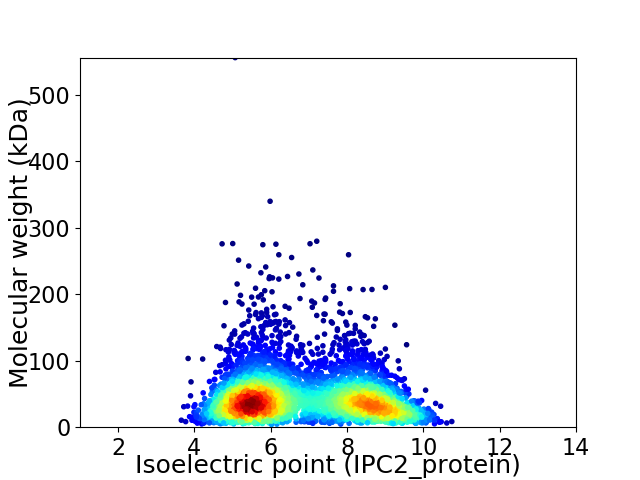
Virtual 2D-PAGE plot for 4024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A656KJQ1|A0A656KJQ1_BLUGR Uncharacterized protein OS=Blumeria graminis f. sp. tritici 96224 OX=1268274 GN=BGT96224_Ac30422 PE=4 SV=1
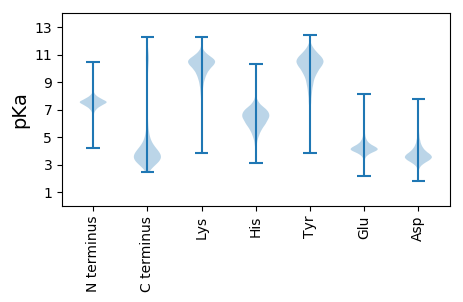
MM1 pKa = 7.83YY2 pKa = 10.17EE3 pKa = 5.34DD4 pKa = 5.75DD5 pKa = 4.46YY6 pKa = 12.09LDD8 pKa = 3.76INEE11 pKa = 4.73TYY13 pKa = 11.21LDD15 pKa = 3.84LFSNEE20 pKa = 3.64TRR22 pKa = 11.84QVSGGVDD29 pKa = 3.44IIGYY33 pKa = 7.44CFPTQPPQPNGYY45 pKa = 8.23GTEE48 pKa = 4.09CLTYY52 pKa = 8.23TGQVSGVFSDD62 pKa = 4.76DD63 pKa = 3.55PSEE66 pKa = 3.91EE67 pKa = 3.92DD68 pKa = 3.36VPRR71 pKa = 11.84KK72 pKa = 9.85RR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.59GIGEE79 pKa = 4.03EE80 pKa = 4.38EE81 pKa = 4.37RR82 pKa = 11.84CDD84 pKa = 3.38EE85 pKa = 4.5DD86 pKa = 4.2RR87 pKa = 11.84AGRR90 pKa = 11.84CEE92 pKa = 3.68
MM1 pKa = 7.83YY2 pKa = 10.17EE3 pKa = 5.34DD4 pKa = 5.75DD5 pKa = 4.46YY6 pKa = 12.09LDD8 pKa = 3.76INEE11 pKa = 4.73TYY13 pKa = 11.21LDD15 pKa = 3.84LFSNEE20 pKa = 3.64TRR22 pKa = 11.84QVSGGVDD29 pKa = 3.44IIGYY33 pKa = 7.44CFPTQPPQPNGYY45 pKa = 8.23GTEE48 pKa = 4.09CLTYY52 pKa = 8.23TGQVSGVFSDD62 pKa = 4.76DD63 pKa = 3.55PSEE66 pKa = 3.91EE67 pKa = 3.92DD68 pKa = 3.36VPRR71 pKa = 11.84KK72 pKa = 9.85RR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.59GIGEE79 pKa = 4.03EE80 pKa = 4.38EE81 pKa = 4.37RR82 pKa = 11.84CDD84 pKa = 3.38EE85 pKa = 4.5DD86 pKa = 4.2RR87 pKa = 11.84AGRR90 pKa = 11.84CEE92 pKa = 3.68
Molecular weight: 10.41 kDa
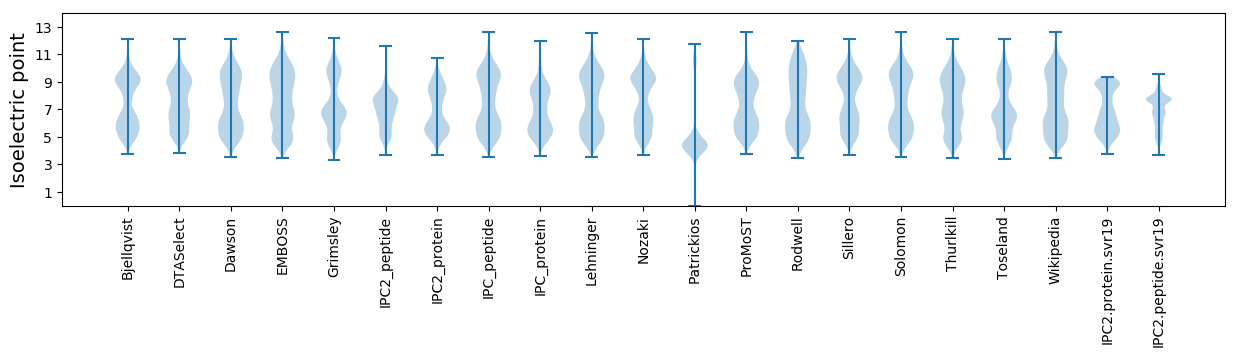
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A656KIX2|A0A656KIX2_BLUGR Uncharacterized protein OS=Blumeria graminis f. sp. tritici 96224 OX=1268274 GN=BGT96224_2832 PE=4 SV=1
MM1 pKa = 7.54GVPPLLSSLAIQGAAKK17 pKa = 10.46AFVHH21 pKa = 5.52GQSRR25 pKa = 11.84ASARR29 pKa = 11.84QSSSDD34 pKa = 3.66SASLTTPALVAARR47 pKa = 11.84ATTVKK52 pKa = 9.5STAKK56 pKa = 10.14NLPASHH62 pKa = 6.74LQPRR66 pKa = 11.84PGDD69 pKa = 3.52APTILGPSRR78 pKa = 11.84PTISTTSHH86 pKa = 5.76VSTPSSRR93 pKa = 11.84SSRR96 pKa = 11.84SSCTQGRR103 pKa = 11.84SAGEE107 pKa = 3.9QASLLYY113 pKa = 10.24RR114 pKa = 11.84PSSLPSSVARR124 pKa = 11.84DD125 pKa = 3.2AAAAAVASTCNLYY138 pKa = 8.41MQPPLSSPCLAPSTITLKK156 pKa = 9.34PTLRR160 pKa = 11.84TSHH163 pKa = 7.04RR164 pKa = 11.84SDD166 pKa = 4.49CEE168 pKa = 3.95DD169 pKa = 3.6SEE171 pKa = 4.57PTCRR175 pKa = 11.84SSAQQRR181 pKa = 11.84RR182 pKa = 11.84SLLHH186 pKa = 6.78PNLHH190 pKa = 4.55QCRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84PMEE199 pKa = 3.85IVTDD203 pKa = 4.12RR204 pKa = 11.84QRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84YY209 pKa = 7.93EE210 pKa = 3.93AVWASNRR217 pKa = 11.84GLYY220 pKa = 10.19LDD222 pKa = 4.23SRR224 pKa = 11.84SHH226 pKa = 5.42EE227 pKa = 3.81AAMRR231 pKa = 11.84LCNLVVRR238 pKa = 11.84DD239 pKa = 3.09IWSRR243 pKa = 11.84SRR245 pKa = 11.84LGSDD249 pKa = 3.3VLHH252 pKa = 7.07CIYY255 pKa = 10.81EE256 pKa = 4.51LIDD259 pKa = 3.88RR260 pKa = 11.84DD261 pKa = 3.52QCGSLRR267 pKa = 11.84RR268 pKa = 11.84DD269 pKa = 3.52EE270 pKa = 5.01FVVGMWLIDD279 pKa = 3.34RR280 pKa = 11.84SLRR283 pKa = 11.84GRR285 pKa = 11.84KK286 pKa = 8.91IPSKK290 pKa = 10.25ISDD293 pKa = 4.47SIWLSVNN300 pKa = 3.07
MM1 pKa = 7.54GVPPLLSSLAIQGAAKK17 pKa = 10.46AFVHH21 pKa = 5.52GQSRR25 pKa = 11.84ASARR29 pKa = 11.84QSSSDD34 pKa = 3.66SASLTTPALVAARR47 pKa = 11.84ATTVKK52 pKa = 9.5STAKK56 pKa = 10.14NLPASHH62 pKa = 6.74LQPRR66 pKa = 11.84PGDD69 pKa = 3.52APTILGPSRR78 pKa = 11.84PTISTTSHH86 pKa = 5.76VSTPSSRR93 pKa = 11.84SSRR96 pKa = 11.84SSCTQGRR103 pKa = 11.84SAGEE107 pKa = 3.9QASLLYY113 pKa = 10.24RR114 pKa = 11.84PSSLPSSVARR124 pKa = 11.84DD125 pKa = 3.2AAAAAVASTCNLYY138 pKa = 8.41MQPPLSSPCLAPSTITLKK156 pKa = 9.34PTLRR160 pKa = 11.84TSHH163 pKa = 7.04RR164 pKa = 11.84SDD166 pKa = 4.49CEE168 pKa = 3.95DD169 pKa = 3.6SEE171 pKa = 4.57PTCRR175 pKa = 11.84SSAQQRR181 pKa = 11.84RR182 pKa = 11.84SLLHH186 pKa = 6.78PNLHH190 pKa = 4.55QCRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84PMEE199 pKa = 3.85IVTDD203 pKa = 4.12RR204 pKa = 11.84QRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84YY209 pKa = 7.93EE210 pKa = 3.93AVWASNRR217 pKa = 11.84GLYY220 pKa = 10.19LDD222 pKa = 4.23SRR224 pKa = 11.84SHH226 pKa = 5.42EE227 pKa = 3.81AAMRR231 pKa = 11.84LCNLVVRR238 pKa = 11.84DD239 pKa = 3.09IWSRR243 pKa = 11.84SRR245 pKa = 11.84LGSDD249 pKa = 3.3VLHH252 pKa = 7.07CIYY255 pKa = 10.81EE256 pKa = 4.51LIDD259 pKa = 3.88RR260 pKa = 11.84DD261 pKa = 3.52QCGSLRR267 pKa = 11.84RR268 pKa = 11.84DD269 pKa = 3.52EE270 pKa = 5.01FVVGMWLIDD279 pKa = 3.34RR280 pKa = 11.84SLRR283 pKa = 11.84GRR285 pKa = 11.84KK286 pKa = 8.91IPSKK290 pKa = 10.25ISDD293 pKa = 4.47SIWLSVNN300 pKa = 3.07
Molecular weight: 32.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1773325 |
36 |
4884 |
440.7 |
49.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.876 ± 0.033 | 1.334 ± 0.012 |
5.408 ± 0.028 | 6.48 ± 0.037 |
3.767 ± 0.02 | 5.79 ± 0.032 |
2.352 ± 0.016 | 6.186 ± 0.025 |
5.955 ± 0.03 | 9.095 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.013 | 4.722 ± 0.021 |
5.313 ± 0.032 | 4.089 ± 0.026 |
5.817 ± 0.026 | 9.243 ± 0.04 |
5.914 ± 0.023 | 5.453 ± 0.025 |
1.229 ± 0.014 | 2.794 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
