
Pararhizobium polonicum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Pararhizobium
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
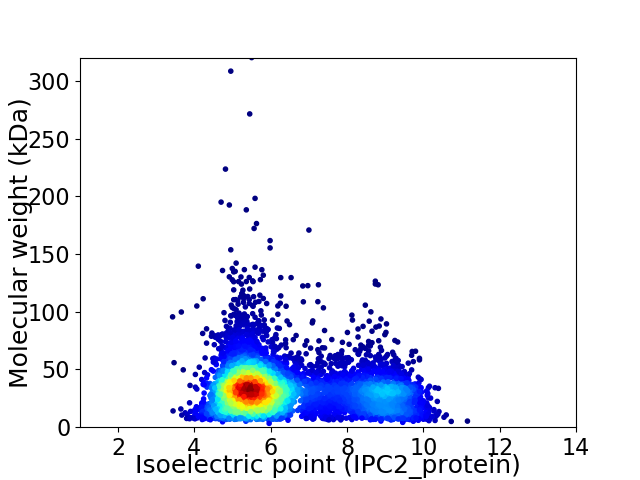
Virtual 2D-PAGE plot for 5658 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C7P209|A0A1C7P209_9RHIZ L-threonine aldolase OS=Pararhizobium polonicum OX=1612624 GN=ADU59_12060 PE=3 SV=1
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 7.48TVLNALGEE11 pKa = 4.35TLFYY15 pKa = 10.75SDD17 pKa = 3.94SSSGFFSATGSGPTLYY33 pKa = 9.14GTAGNDD39 pKa = 4.03SMWGDD44 pKa = 3.58SAVNVTMSGGTGDD57 pKa = 4.91DD58 pKa = 2.72IYY60 pKa = 11.64YY61 pKa = 10.28LYY63 pKa = 10.97SAINRR68 pKa = 11.84AYY70 pKa = 8.75EE71 pKa = 3.86AAGEE75 pKa = 4.54GIDD78 pKa = 5.25TISTWMSYY86 pKa = 7.29TLPDD90 pKa = 3.39NFEE93 pKa = 4.16NLIVTGDD100 pKa = 3.47LRR102 pKa = 11.84HH103 pKa = 6.38AFGNSADD110 pKa = 4.01NIITGGTGRR119 pKa = 11.84QTIDD123 pKa = 2.81GGAGNDD129 pKa = 3.45VLTGGGGADD138 pKa = 3.05TFIFSKK144 pKa = 11.18GNGSDD149 pKa = 4.93LITDD153 pKa = 4.45FAADD157 pKa = 3.4DD158 pKa = 3.97TVRR161 pKa = 11.84LNGYY165 pKa = 9.12SLTSFDD171 pKa = 4.0QLLSNTTQEE180 pKa = 4.4GANLRR185 pKa = 11.84LDD187 pKa = 4.09LGDD190 pKa = 4.17GEE192 pKa = 4.98SLVFADD198 pKa = 4.55TTASDD203 pKa = 3.79LNAGQFEE210 pKa = 4.57LSLDD214 pKa = 3.7RR215 pKa = 11.84SALTQTFNDD224 pKa = 3.58NFDD227 pKa = 3.79TLSLVNGTSGVWEE240 pKa = 4.25AKK242 pKa = 10.05YY243 pKa = 9.18WWAPDD248 pKa = 3.07KK249 pKa = 11.31GATLTEE255 pKa = 4.14NGEE258 pKa = 4.27LQWYY262 pKa = 7.8VNPLYY267 pKa = 10.95APTSAVNPFSIDD279 pKa = 3.35NGVLTITAEE288 pKa = 3.92QTPASISAEE297 pKa = 4.05VEE299 pKa = 4.33GYY301 pKa = 10.77DD302 pKa = 3.71YY303 pKa = 11.14TSGMLTTHH311 pKa = 7.26ASFAQTYY318 pKa = 9.42GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.63MPDD329 pKa = 4.41DD330 pKa = 4.11KK331 pKa = 11.4GVWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.78LDD351 pKa = 3.52VVEE354 pKa = 5.06MRR356 pKa = 11.84GQDD359 pKa = 3.55PNTVHH364 pKa = 5.48VTVHH368 pKa = 5.78SNEE371 pKa = 4.45TGSQTSVSSAVSVSDD386 pKa = 3.24TEE388 pKa = 4.82GFHH391 pKa = 6.5TYY393 pKa = 9.81GVLWQEE399 pKa = 5.31DD400 pKa = 4.39EE401 pKa = 4.4IVWYY405 pKa = 9.99IDD407 pKa = 3.26DD408 pKa = 4.0VAVARR413 pKa = 11.84ADD415 pKa = 3.9TPSDD419 pKa = 3.23MHH421 pKa = 7.22EE422 pKa = 4.05PMYY425 pKa = 10.32MLVNLAVGGMAGTPTDD441 pKa = 3.87GLPDD445 pKa = 3.95GSEE448 pKa = 3.71MKK450 pKa = 9.8IDD452 pKa = 3.92YY453 pKa = 10.28IRR455 pKa = 11.84AYY457 pKa = 9.6TLDD460 pKa = 4.0DD461 pKa = 3.86LAA463 pKa = 6.68
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 7.48TVLNALGEE11 pKa = 4.35TLFYY15 pKa = 10.75SDD17 pKa = 3.94SSSGFFSATGSGPTLYY33 pKa = 9.14GTAGNDD39 pKa = 4.03SMWGDD44 pKa = 3.58SAVNVTMSGGTGDD57 pKa = 4.91DD58 pKa = 2.72IYY60 pKa = 11.64YY61 pKa = 10.28LYY63 pKa = 10.97SAINRR68 pKa = 11.84AYY70 pKa = 8.75EE71 pKa = 3.86AAGEE75 pKa = 4.54GIDD78 pKa = 5.25TISTWMSYY86 pKa = 7.29TLPDD90 pKa = 3.39NFEE93 pKa = 4.16NLIVTGDD100 pKa = 3.47LRR102 pKa = 11.84HH103 pKa = 6.38AFGNSADD110 pKa = 4.01NIITGGTGRR119 pKa = 11.84QTIDD123 pKa = 2.81GGAGNDD129 pKa = 3.45VLTGGGGADD138 pKa = 3.05TFIFSKK144 pKa = 11.18GNGSDD149 pKa = 4.93LITDD153 pKa = 4.45FAADD157 pKa = 3.4DD158 pKa = 3.97TVRR161 pKa = 11.84LNGYY165 pKa = 9.12SLTSFDD171 pKa = 4.0QLLSNTTQEE180 pKa = 4.4GANLRR185 pKa = 11.84LDD187 pKa = 4.09LGDD190 pKa = 4.17GEE192 pKa = 4.98SLVFADD198 pKa = 4.55TTASDD203 pKa = 3.79LNAGQFEE210 pKa = 4.57LSLDD214 pKa = 3.7RR215 pKa = 11.84SALTQTFNDD224 pKa = 3.58NFDD227 pKa = 3.79TLSLVNGTSGVWEE240 pKa = 4.25AKK242 pKa = 10.05YY243 pKa = 9.18WWAPDD248 pKa = 3.07KK249 pKa = 11.31GATLTEE255 pKa = 4.14NGEE258 pKa = 4.27LQWYY262 pKa = 7.8VNPLYY267 pKa = 10.95APTSAVNPFSIDD279 pKa = 3.35NGVLTITAEE288 pKa = 3.92QTPASISAEE297 pKa = 4.05VEE299 pKa = 4.33GYY301 pKa = 10.77DD302 pKa = 3.71YY303 pKa = 11.14TSGMLTTHH311 pKa = 7.26ASFAQTYY318 pKa = 9.42GYY320 pKa = 10.77FEE322 pKa = 4.72IRR324 pKa = 11.84ADD326 pKa = 3.63MPDD329 pKa = 4.41DD330 pKa = 4.11KK331 pKa = 11.4GVWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.78LDD351 pKa = 3.52VVEE354 pKa = 5.06MRR356 pKa = 11.84GQDD359 pKa = 3.55PNTVHH364 pKa = 5.48VTVHH368 pKa = 5.78SNEE371 pKa = 4.45TGSQTSVSSAVSVSDD386 pKa = 3.24TEE388 pKa = 4.82GFHH391 pKa = 6.5TYY393 pKa = 9.81GVLWQEE399 pKa = 5.31DD400 pKa = 4.39EE401 pKa = 4.4IVWYY405 pKa = 9.99IDD407 pKa = 3.26DD408 pKa = 4.0VAVARR413 pKa = 11.84ADD415 pKa = 3.9TPSDD419 pKa = 3.23MHH421 pKa = 7.22EE422 pKa = 4.05PMYY425 pKa = 10.32MLVNLAVGGMAGTPTDD441 pKa = 3.87GLPDD445 pKa = 3.95GSEE448 pKa = 3.71MKK450 pKa = 9.8IDD452 pKa = 3.92YY453 pKa = 10.28IRR455 pKa = 11.84AYY457 pKa = 9.6TLDD460 pKa = 4.0DD461 pKa = 3.86LAA463 pKa = 6.68
Molecular weight: 49.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C7P6G5|A0A1C7P6G5_9RHIZ Nodulation protein N OS=Pararhizobium polonicum OX=1612624 GN=ADU59_11890 PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.64GFRR20 pKa = 11.84ARR22 pKa = 11.84IATKK26 pKa = 9.98GGRR29 pKa = 11.84KK30 pKa = 9.73VIVARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1784372 |
29 |
2875 |
315.4 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.91 ± 0.039 | 0.783 ± 0.011 |
5.715 ± 0.024 | 5.521 ± 0.032 |
3.989 ± 0.02 | 8.465 ± 0.029 |
1.995 ± 0.015 | 5.876 ± 0.025 |
3.72 ± 0.025 | 10.026 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.601 ± 0.015 | 2.852 ± 0.018 |
4.846 ± 0.019 | 3.113 ± 0.019 |
6.408 ± 0.028 | 5.763 ± 0.024 |
5.536 ± 0.02 | 7.309 ± 0.024 |
1.255 ± 0.012 | 2.319 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
