
Enhydra lutris papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Lambdapapillomavirus; Lambdapapillomavirus 4
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
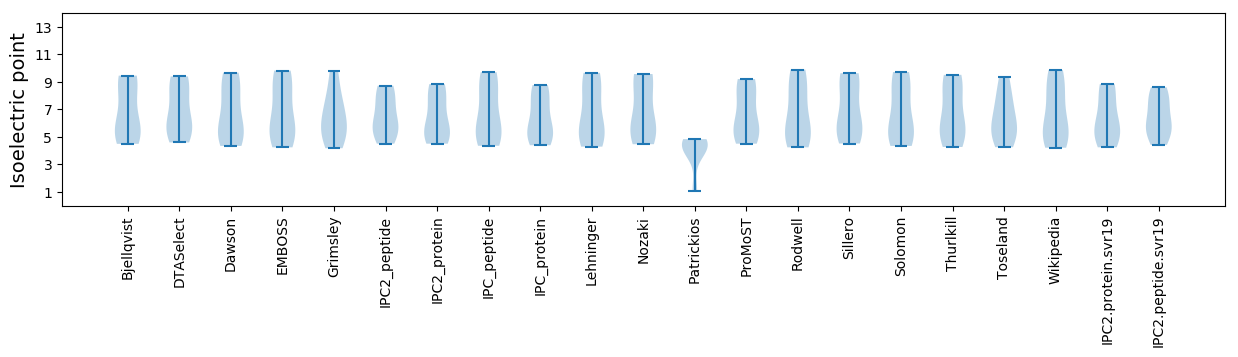
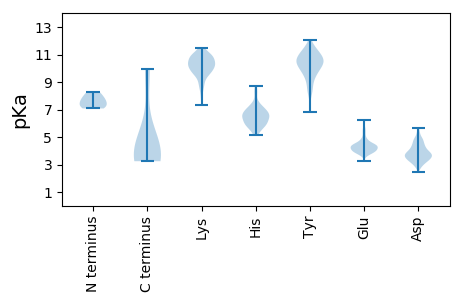
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8R555|W8R555_9PAPI Major capsid protein L1 OS=Enhydra lutris papillomavirus 1 OX=1472717 GN=L1 PE=3 SV=1
MM1 pKa = 7.62FGLCPTLGEE10 pKa = 4.4VVLTEE15 pKa = 3.86QPEE18 pKa = 4.5AIDD21 pKa = 3.39LHH23 pKa = 8.68CYY25 pKa = 8.29EE26 pKa = 5.32HH27 pKa = 6.65MPSDD31 pKa = 3.89DD32 pKa = 4.03EE33 pKa = 4.51EE34 pKa = 4.99EE35 pKa = 4.37EE36 pKa = 4.3EE37 pKa = 4.17QTSRR41 pKa = 11.84DD42 pKa = 3.7LYY44 pKa = 10.61RR45 pKa = 11.84VSVDD49 pKa = 3.09CGACKK54 pKa = 9.61RR55 pKa = 11.84TVTLVVFADD64 pKa = 4.02LEE66 pKa = 4.92DD67 pKa = 3.68IQKK70 pKa = 10.08LHH72 pKa = 6.64SLLHH76 pKa = 5.14SVRR79 pKa = 11.84VVCEE83 pKa = 3.77SCLNPNNLNHH93 pKa = 6.88GGG95 pKa = 3.53
MM1 pKa = 7.62FGLCPTLGEE10 pKa = 4.4VVLTEE15 pKa = 3.86QPEE18 pKa = 4.5AIDD21 pKa = 3.39LHH23 pKa = 8.68CYY25 pKa = 8.29EE26 pKa = 5.32HH27 pKa = 6.65MPSDD31 pKa = 3.89DD32 pKa = 4.03EE33 pKa = 4.51EE34 pKa = 4.99EE35 pKa = 4.37EE36 pKa = 4.3EE37 pKa = 4.17QTSRR41 pKa = 11.84DD42 pKa = 3.7LYY44 pKa = 10.61RR45 pKa = 11.84VSVDD49 pKa = 3.09CGACKK54 pKa = 9.61RR55 pKa = 11.84TVTLVVFADD64 pKa = 4.02LEE66 pKa = 4.92DD67 pKa = 3.68IQKK70 pKa = 10.08LHH72 pKa = 6.64SLLHH76 pKa = 5.14SVRR79 pKa = 11.84VVCEE83 pKa = 3.77SCLNPNNLNHH93 pKa = 6.88GGG95 pKa = 3.53
Molecular weight: 10.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8S105|W8S105_9PAPI Replication protein E1 OS=Enhydra lutris papillomavirus 1 OX=1472717 GN=E1 PE=3 SV=1
MM1 pKa = 7.08EE2 pKa = 5.13TLNQALNSVQEE13 pKa = 4.54DD14 pKa = 4.05LLSLYY19 pKa = 8.41EE20 pKa = 4.12QNSEE24 pKa = 4.76SITDD28 pKa = 4.7QIQHH32 pKa = 5.95WNLLRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.01QVILYY44 pKa = 6.84YY45 pKa = 10.69ARR47 pKa = 11.84EE48 pKa = 3.9RR49 pKa = 11.84GITRR53 pKa = 11.84LGMTLVPPRR62 pKa = 11.84HH63 pKa = 5.97VSQQKK68 pKa = 9.8AKK70 pKa = 10.53AAIEE74 pKa = 3.89QVLYY78 pKa = 9.18LTSLSQSSFKK88 pKa = 10.71DD89 pKa = 3.63EE90 pKa = 3.87RR91 pKa = 11.84WTLGDD96 pKa = 3.31TSRR99 pKa = 11.84EE100 pKa = 3.9RR101 pKa = 11.84FLTEE105 pKa = 4.38PEE107 pKa = 3.84NCFKK111 pKa = 11.05KK112 pKa = 10.51GGQQIDD118 pKa = 3.52VKK120 pKa = 11.37YY121 pKa = 10.72GGEE124 pKa = 3.66EE125 pKa = 4.1DD126 pKa = 3.92NIVRR130 pKa = 11.84YY131 pKa = 7.1TLWANIYY138 pKa = 8.45FQNEE142 pKa = 3.29QDD144 pKa = 3.29SWEE147 pKa = 4.12KK148 pKa = 11.5AEE150 pKa = 4.59GKK152 pKa = 10.06VDD154 pKa = 3.81NKK156 pKa = 10.77GLYY159 pKa = 10.0YY160 pKa = 10.63EE161 pKa = 5.24DD162 pKa = 4.46ADD164 pKa = 4.04GVRR167 pKa = 11.84TYY169 pKa = 11.24YY170 pKa = 11.02VDD172 pKa = 4.86FIKK175 pKa = 10.67EE176 pKa = 3.87ATKK179 pKa = 10.8YY180 pKa = 10.57SSNGQYY186 pKa = 10.44EE187 pKa = 4.25VLQRR191 pKa = 11.84VNPVSRR197 pKa = 11.84PASGAVNSAPLGCASHH213 pKa = 7.24CSTPKK218 pKa = 10.49KK219 pKa = 9.35KK220 pKa = 8.68TADD223 pKa = 3.31PPRR226 pKa = 11.84RR227 pKa = 11.84GNYY230 pKa = 8.37RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84SKK236 pKa = 10.31PSKK239 pKa = 9.99QGQHH243 pKa = 5.84SPGGRR248 pKa = 11.84QGKK251 pKa = 8.09QASTSRR257 pKa = 11.84PTAPSPGEE265 pKa = 3.67VGRR268 pKa = 11.84SHH270 pKa = 7.37KK271 pKa = 10.47SASPRR276 pKa = 11.84LGGRR280 pKa = 11.84LEE282 pKa = 4.32RR283 pKa = 11.84LIQEE287 pKa = 4.43ARR289 pKa = 11.84DD290 pKa = 3.64PPIVVLKK297 pKa = 11.2GEE299 pKa = 4.4ANPLKK304 pKa = 10.11CLRR307 pKa = 11.84YY308 pKa = 9.55RR309 pKa = 11.84LTKK312 pKa = 10.24GYY314 pKa = 10.49SSTFEE319 pKa = 4.11EE320 pKa = 6.21VSTTWKK326 pKa = 9.3WASCKK331 pKa = 10.12TKK333 pKa = 10.52DD334 pKa = 3.26PCGRR338 pKa = 11.84ARR340 pKa = 11.84MLVSFSTPDD349 pKa = 3.18QRR351 pKa = 11.84DD352 pKa = 3.11LFLQHH357 pKa = 6.24VPLPKK362 pKa = 9.64SVKK365 pKa = 10.26YY366 pKa = 9.74FLGSLNDD373 pKa = 3.34II374 pKa = 4.77
MM1 pKa = 7.08EE2 pKa = 5.13TLNQALNSVQEE13 pKa = 4.54DD14 pKa = 4.05LLSLYY19 pKa = 8.41EE20 pKa = 4.12QNSEE24 pKa = 4.76SITDD28 pKa = 4.7QIQHH32 pKa = 5.95WNLLRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.01QVILYY44 pKa = 6.84YY45 pKa = 10.69ARR47 pKa = 11.84EE48 pKa = 3.9RR49 pKa = 11.84GITRR53 pKa = 11.84LGMTLVPPRR62 pKa = 11.84HH63 pKa = 5.97VSQQKK68 pKa = 9.8AKK70 pKa = 10.53AAIEE74 pKa = 3.89QVLYY78 pKa = 9.18LTSLSQSSFKK88 pKa = 10.71DD89 pKa = 3.63EE90 pKa = 3.87RR91 pKa = 11.84WTLGDD96 pKa = 3.31TSRR99 pKa = 11.84EE100 pKa = 3.9RR101 pKa = 11.84FLTEE105 pKa = 4.38PEE107 pKa = 3.84NCFKK111 pKa = 11.05KK112 pKa = 10.51GGQQIDD118 pKa = 3.52VKK120 pKa = 11.37YY121 pKa = 10.72GGEE124 pKa = 3.66EE125 pKa = 4.1DD126 pKa = 3.92NIVRR130 pKa = 11.84YY131 pKa = 7.1TLWANIYY138 pKa = 8.45FQNEE142 pKa = 3.29QDD144 pKa = 3.29SWEE147 pKa = 4.12KK148 pKa = 11.5AEE150 pKa = 4.59GKK152 pKa = 10.06VDD154 pKa = 3.81NKK156 pKa = 10.77GLYY159 pKa = 10.0YY160 pKa = 10.63EE161 pKa = 5.24DD162 pKa = 4.46ADD164 pKa = 4.04GVRR167 pKa = 11.84TYY169 pKa = 11.24YY170 pKa = 11.02VDD172 pKa = 4.86FIKK175 pKa = 10.67EE176 pKa = 3.87ATKK179 pKa = 10.8YY180 pKa = 10.57SSNGQYY186 pKa = 10.44EE187 pKa = 4.25VLQRR191 pKa = 11.84VNPVSRR197 pKa = 11.84PASGAVNSAPLGCASHH213 pKa = 7.24CSTPKK218 pKa = 10.49KK219 pKa = 9.35KK220 pKa = 8.68TADD223 pKa = 3.31PPRR226 pKa = 11.84RR227 pKa = 11.84GNYY230 pKa = 8.37RR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84SKK236 pKa = 10.31PSKK239 pKa = 9.99QGQHH243 pKa = 5.84SPGGRR248 pKa = 11.84QGKK251 pKa = 8.09QASTSRR257 pKa = 11.84PTAPSPGEE265 pKa = 3.67VGRR268 pKa = 11.84SHH270 pKa = 7.37KK271 pKa = 10.47SASPRR276 pKa = 11.84LGGRR280 pKa = 11.84LEE282 pKa = 4.32RR283 pKa = 11.84LIQEE287 pKa = 4.43ARR289 pKa = 11.84DD290 pKa = 3.64PPIVVLKK297 pKa = 11.2GEE299 pKa = 4.4ANPLKK304 pKa = 10.11CLRR307 pKa = 11.84YY308 pKa = 9.55RR309 pKa = 11.84LTKK312 pKa = 10.24GYY314 pKa = 10.49SSTFEE319 pKa = 4.11EE320 pKa = 6.21VSTTWKK326 pKa = 9.3WASCKK331 pKa = 10.12TKK333 pKa = 10.52DD334 pKa = 3.26PCGRR338 pKa = 11.84ARR340 pKa = 11.84MLVSFSTPDD349 pKa = 3.18QRR351 pKa = 11.84DD352 pKa = 3.11LFLQHH357 pKa = 6.24VPLPKK362 pKa = 9.64SVKK365 pKa = 10.26YY366 pKa = 9.74FLGSLNDD373 pKa = 3.34II374 pKa = 4.77
Molecular weight: 42.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2335 |
95 |
596 |
333.6 |
37.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.91 ± 0.422 | 2.313 ± 0.846 |
6.253 ± 0.41 | 6.039 ± 0.486 |
4.625 ± 0.562 | 6.124 ± 0.662 |
2.184 ± 0.185 | 3.897 ± 0.689 |
5.61 ± 0.712 | 9.208 ± 0.886 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.413 ± 0.324 | 4.197 ± 0.667 |
6.51 ± 1.099 | 4.754 ± 0.374 |
6.124 ± 0.694 | 8.051 ± 0.689 |
6.253 ± 0.489 | 6.296 ± 0.812 |
1.285 ± 0.392 | 2.955 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
