
Halarchaeum sp. CBA1220
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halarchaeum; unclassified Halarchaeum
Average proteome isoelectric point is 4.92
Get precalculated fractions of proteins
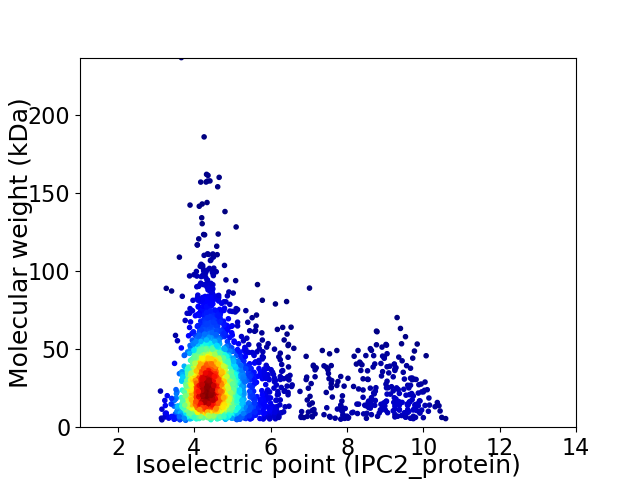
Virtual 2D-PAGE plot for 3110 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
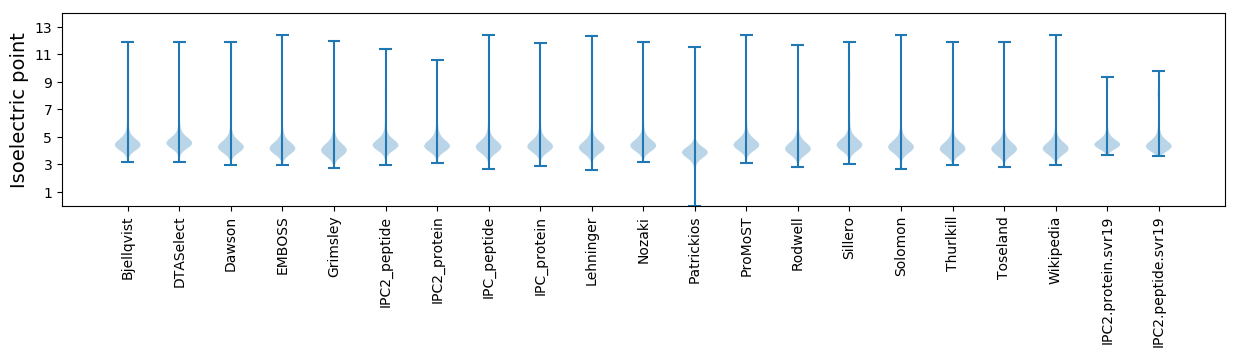
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7D5I613|A0A7D5I613_9EURY PH domain-containing protein OS=Halarchaeum sp. CBA1220 OX=1853682 GN=EFA46_012190 PE=4 SV=1
MM1 pKa = 7.52SNFDD5 pKa = 3.28RR6 pKa = 11.84RR7 pKa = 11.84DD8 pKa = 3.54FVKK11 pKa = 9.64VTGGLGVAGLTGLAGCSGGPSGDD34 pKa = 3.45GGGSEE39 pKa = 4.44TTTTSGEE46 pKa = 4.24TSDD49 pKa = 5.67GEE51 pKa = 4.6TTTEE55 pKa = 4.01AEE57 pKa = 3.81AAANIGLVYY66 pKa = 9.58GTGGTGDD73 pKa = 3.66GSFNDD78 pKa = 3.6QAKK81 pKa = 10.26QGIEE85 pKa = 3.91QAVEE89 pKa = 3.95EE90 pKa = 4.7LDD92 pKa = 3.18IAYY95 pKa = 10.29QSAEE99 pKa = 4.12PDD101 pKa = 3.33EE102 pKa = 4.85VSQFSNYY109 pKa = 7.14QQQFASSTDD118 pKa = 3.4PNYY121 pKa = 11.3DD122 pKa = 3.27LVSCIGYY129 pKa = 8.98LQADD133 pKa = 4.33ALSEE137 pKa = 4.32SASQYY142 pKa = 10.45PDD144 pKa = 3.11QQFQIVDD151 pKa = 3.75STVEE155 pKa = 3.85GDD157 pKa = 3.5NVASYY162 pKa = 10.86VFQEE166 pKa = 4.36HH167 pKa = 6.52EE168 pKa = 4.48GSFLTGVLAGHH179 pKa = 7.37LTTMSFEE186 pKa = 4.45AGAGATQSDD195 pKa = 4.24STNVGFVGGVEE206 pKa = 4.01GDD208 pKa = 4.15LIGKK212 pKa = 9.19FEE214 pKa = 4.76AGYY217 pKa = 10.08KK218 pKa = 10.47AGVKK222 pKa = 9.45YY223 pKa = 11.04ANDD226 pKa = 3.62DD227 pKa = 3.32VDD229 pKa = 3.39IQTSYY234 pKa = 11.22VGSFNDD240 pKa = 3.56PSGGQEE246 pKa = 3.9AALAMYY252 pKa = 10.6NSGADD257 pKa = 3.19IVYY260 pKa = 9.58HH261 pKa = 6.5ASGNTGTGVFQAAQQKK277 pKa = 9.3GRR279 pKa = 11.84FAIGVDD285 pKa = 3.54RR286 pKa = 11.84DD287 pKa = 3.49QSLTKK292 pKa = 10.05PSYY295 pKa = 11.23ADD297 pKa = 3.45VILASMVKK305 pKa = 10.16RR306 pKa = 11.84VDD308 pKa = 3.32TAVYY312 pKa = 9.15NCVKK316 pKa = 10.7NVVNEE321 pKa = 3.93NFNGGSTTTLGLQDD335 pKa = 3.85GGVACVYY342 pKa = 10.71GDD344 pKa = 4.06EE345 pKa = 4.73LGGDD349 pKa = 3.18IPEE352 pKa = 4.46DD353 pKa = 3.37VKK355 pKa = 10.64TAVADD360 pKa = 3.75ARR362 pKa = 11.84QSIIDD367 pKa = 3.71GDD369 pKa = 3.69IDD371 pKa = 4.66VPTDD375 pKa = 3.6PNDD378 pKa = 3.18AA379 pKa = 4.24
MM1 pKa = 7.52SNFDD5 pKa = 3.28RR6 pKa = 11.84RR7 pKa = 11.84DD8 pKa = 3.54FVKK11 pKa = 9.64VTGGLGVAGLTGLAGCSGGPSGDD34 pKa = 3.45GGGSEE39 pKa = 4.44TTTTSGEE46 pKa = 4.24TSDD49 pKa = 5.67GEE51 pKa = 4.6TTTEE55 pKa = 4.01AEE57 pKa = 3.81AAANIGLVYY66 pKa = 9.58GTGGTGDD73 pKa = 3.66GSFNDD78 pKa = 3.6QAKK81 pKa = 10.26QGIEE85 pKa = 3.91QAVEE89 pKa = 3.95EE90 pKa = 4.7LDD92 pKa = 3.18IAYY95 pKa = 10.29QSAEE99 pKa = 4.12PDD101 pKa = 3.33EE102 pKa = 4.85VSQFSNYY109 pKa = 7.14QQQFASSTDD118 pKa = 3.4PNYY121 pKa = 11.3DD122 pKa = 3.27LVSCIGYY129 pKa = 8.98LQADD133 pKa = 4.33ALSEE137 pKa = 4.32SASQYY142 pKa = 10.45PDD144 pKa = 3.11QQFQIVDD151 pKa = 3.75STVEE155 pKa = 3.85GDD157 pKa = 3.5NVASYY162 pKa = 10.86VFQEE166 pKa = 4.36HH167 pKa = 6.52EE168 pKa = 4.48GSFLTGVLAGHH179 pKa = 7.37LTTMSFEE186 pKa = 4.45AGAGATQSDD195 pKa = 4.24STNVGFVGGVEE206 pKa = 4.01GDD208 pKa = 4.15LIGKK212 pKa = 9.19FEE214 pKa = 4.76AGYY217 pKa = 10.08KK218 pKa = 10.47AGVKK222 pKa = 9.45YY223 pKa = 11.04ANDD226 pKa = 3.62DD227 pKa = 3.32VDD229 pKa = 3.39IQTSYY234 pKa = 11.22VGSFNDD240 pKa = 3.56PSGGQEE246 pKa = 3.9AALAMYY252 pKa = 10.6NSGADD257 pKa = 3.19IVYY260 pKa = 9.58HH261 pKa = 6.5ASGNTGTGVFQAAQQKK277 pKa = 9.3GRR279 pKa = 11.84FAIGVDD285 pKa = 3.54RR286 pKa = 11.84DD287 pKa = 3.49QSLTKK292 pKa = 10.05PSYY295 pKa = 11.23ADD297 pKa = 3.45VILASMVKK305 pKa = 10.16RR306 pKa = 11.84VDD308 pKa = 3.32TAVYY312 pKa = 9.15NCVKK316 pKa = 10.7NVVNEE321 pKa = 3.93NFNGGSTTTLGLQDD335 pKa = 3.85GGVACVYY342 pKa = 10.71GDD344 pKa = 4.06EE345 pKa = 4.73LGGDD349 pKa = 3.18IPEE352 pKa = 4.46DD353 pKa = 3.37VKK355 pKa = 10.64TAVADD360 pKa = 3.75ARR362 pKa = 11.84QSIIDD367 pKa = 3.71GDD369 pKa = 3.69IDD371 pKa = 4.66VPTDD375 pKa = 3.6PNDD378 pKa = 3.18AA379 pKa = 4.24
Molecular weight: 39.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7D5E1M4|A0A7D5E1M4_9EURY Cupin domain-containing protein OS=Halarchaeum sp. CBA1220 OX=1853682 GN=EFA46_001720 PE=4 SV=1
MM1 pKa = 8.05RR2 pKa = 11.84PGLKK6 pKa = 10.0VGSWTFGLTALLVVTVLVSAGIGTVSIPALTVARR40 pKa = 11.84VCLNALVVPTSLSVHH55 pKa = 5.89TSALGSTGVGLPTVSVGTTHH75 pKa = 7.52LFDD78 pKa = 3.99FRR80 pKa = 11.84TPEE83 pKa = 3.66TSRR86 pKa = 11.84IIVMTIRR93 pKa = 11.84LPRR96 pKa = 11.84IALASLVGFALALAGTVMQGFFRR119 pKa = 11.84NPMADD124 pKa = 3.31PSIIGVSSGAAVGAVTVITVGVSFPFGLGIHH155 pKa = 6.53GAAFLGGLLAAFGIYY170 pKa = 10.6ALGTRR175 pKa = 11.84GGRR178 pKa = 11.84TPVTTLLLAGVAVEE192 pKa = 4.32TFLGAVISYY201 pKa = 8.63MLVQSGNEE209 pKa = 3.85LRR211 pKa = 11.84AAMYY215 pKa = 9.89WLMGHH220 pKa = 6.79LQNASWGDD228 pKa = 3.53VTATALTLPLLFGVLLGYY246 pKa = 9.91RR247 pKa = 11.84RR248 pKa = 11.84HH249 pKa = 6.49LNVLLLGEE257 pKa = 4.91HH258 pKa = 6.93DD259 pKa = 4.4AKK261 pKa = 10.55TLGIEE266 pKa = 4.01VEE268 pKa = 4.06RR269 pKa = 11.84TKK271 pKa = 10.73RR272 pKa = 11.84ILLAVSALVTAIAVAVSGVIGFVGLVVPHH301 pKa = 6.13VMRR304 pKa = 11.84LLVGPDD310 pKa = 3.3HH311 pKa = 7.45RR312 pKa = 11.84ILLPTSALAGASFLVAADD330 pKa = 3.91TVARR334 pKa = 11.84SGPAQLPVGIVTAFVGAPFFLYY356 pKa = 10.72LLRR359 pKa = 11.84KK360 pKa = 9.95RR361 pKa = 11.84EE362 pKa = 4.0VAEE365 pKa = 4.03LL366 pKa = 3.58
MM1 pKa = 8.05RR2 pKa = 11.84PGLKK6 pKa = 10.0VGSWTFGLTALLVVTVLVSAGIGTVSIPALTVARR40 pKa = 11.84VCLNALVVPTSLSVHH55 pKa = 5.89TSALGSTGVGLPTVSVGTTHH75 pKa = 7.52LFDD78 pKa = 3.99FRR80 pKa = 11.84TPEE83 pKa = 3.66TSRR86 pKa = 11.84IIVMTIRR93 pKa = 11.84LPRR96 pKa = 11.84IALASLVGFALALAGTVMQGFFRR119 pKa = 11.84NPMADD124 pKa = 3.31PSIIGVSSGAAVGAVTVITVGVSFPFGLGIHH155 pKa = 6.53GAAFLGGLLAAFGIYY170 pKa = 10.6ALGTRR175 pKa = 11.84GGRR178 pKa = 11.84TPVTTLLLAGVAVEE192 pKa = 4.32TFLGAVISYY201 pKa = 8.63MLVQSGNEE209 pKa = 3.85LRR211 pKa = 11.84AAMYY215 pKa = 9.89WLMGHH220 pKa = 6.79LQNASWGDD228 pKa = 3.53VTATALTLPLLFGVLLGYY246 pKa = 9.91RR247 pKa = 11.84RR248 pKa = 11.84HH249 pKa = 6.49LNVLLLGEE257 pKa = 4.91HH258 pKa = 6.93DD259 pKa = 4.4AKK261 pKa = 10.55TLGIEE266 pKa = 4.01VEE268 pKa = 4.06RR269 pKa = 11.84TKK271 pKa = 10.73RR272 pKa = 11.84ILLAVSALVTAIAVAVSGVIGFVGLVVPHH301 pKa = 6.13VMRR304 pKa = 11.84LLVGPDD310 pKa = 3.3HH311 pKa = 7.45RR312 pKa = 11.84ILLPTSALAGASFLVAADD330 pKa = 3.91TVARR334 pKa = 11.84SGPAQLPVGIVTAFVGAPFFLYY356 pKa = 10.72LLRR359 pKa = 11.84KK360 pKa = 9.95RR361 pKa = 11.84EE362 pKa = 4.0VAEE365 pKa = 4.03LL366 pKa = 3.58
Molecular weight: 37.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
920609 |
44 |
2329 |
296.0 |
32.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.249 ± 0.096 | 0.724 ± 0.014 |
8.434 ± 0.057 | 8.306 ± 0.072 |
3.163 ± 0.03 | 8.463 ± 0.049 |
2.123 ± 0.022 | 3.191 ± 0.04 |
1.724 ± 0.03 | 9.11 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.605 ± 0.021 | 2.194 ± 0.028 |
4.588 ± 0.029 | 2.195 ± 0.026 |
6.779 ± 0.046 | 5.211 ± 0.037 |
6.315 ± 0.049 | 9.666 ± 0.053 |
1.131 ± 0.017 | 2.83 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
