
Granulicella mallensis (strain ATCC BAA-1857 / DSM 23137 / MP5ACTX8)
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Granulicella; Granulicella mallensis
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
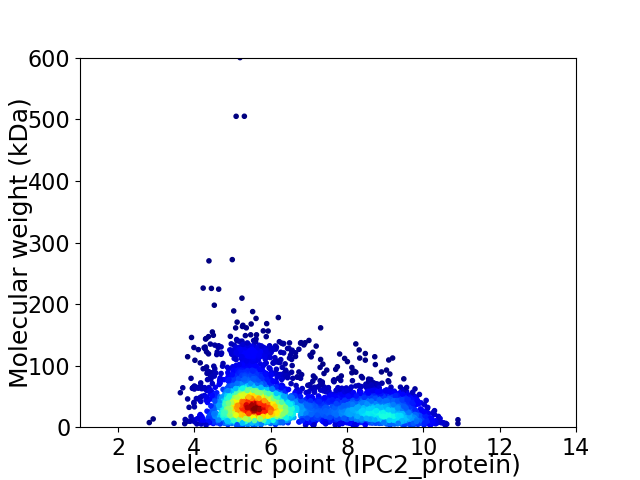
Virtual 2D-PAGE plot for 4804 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8NRU2|G8NRU2_GRAMM Cna B-type protein OS=Granulicella mallensis (strain ATCC BAA-1857 / DSM 23137 / MP5ACTX8) OX=682795 GN=AciX8_0706 PE=4 SV=1
MM1 pKa = 7.09QLYY4 pKa = 10.37AAGTSGNGSAPQALLSSAVTTDD26 pKa = 2.91ATGSFTVPSGYY37 pKa = 8.65TCPSATAITYY47 pKa = 10.03LLAEE51 pKa = 5.28GGQAAGASSANADD64 pKa = 3.92LWLMAPIGACGDD76 pKa = 3.29ITASTTAVLNEE87 pKa = 3.97VTTVASAWALSQFLAPEE104 pKa = 4.62GNLGATSTNALGLANAVIVANNLVNIATGTSPGAAVPANVTVSTAKK150 pKa = 10.84LNTLADD156 pKa = 3.93ALSSCMAPAGEE167 pKa = 4.27TACAALLSAATTSGTTPGNTLDD189 pKa = 3.64AALNIARR196 pKa = 11.84HH197 pKa = 5.47PANNVSSIYY206 pKa = 10.46TLAQANAIFPSLTAAPPDD224 pKa = 3.22WLLYY228 pKa = 9.3RR229 pKa = 11.84TITGGGLEE237 pKa = 4.01QTPRR241 pKa = 11.84AIAIDD246 pKa = 3.48SAGNVWVATDD256 pKa = 3.41YY257 pKa = 11.55GAVAKK262 pKa = 10.41FSTNGTPVAPEE273 pKa = 4.28GFTGTGVNEE282 pKa = 4.28SRR284 pKa = 11.84SMALDD289 pKa = 3.06VHH291 pKa = 6.57DD292 pKa = 5.01NVWIANRR299 pKa = 11.84EE300 pKa = 4.06TTQNSGLGDD309 pKa = 4.09VIEE312 pKa = 4.85LSPTGQLLSGTGGFTDD328 pKa = 4.28GGINFPQGIATDD340 pKa = 4.03PNGNVWVANANLSTVTLLNDD360 pKa = 3.36SGTVLAASIGAGGGLIAVPDD380 pKa = 5.39AIAVDD385 pKa = 3.97ANDD388 pKa = 3.78NAWVGNTGAGSTITRR403 pKa = 11.84ISSDD407 pKa = 2.8GSQVTNLTCCDD418 pKa = 3.84HH419 pKa = 7.06PAGIAVDD426 pKa = 3.7QEE428 pKa = 4.79DD429 pKa = 4.45NVWTANFLDD438 pKa = 5.05DD439 pKa = 5.23SITMITNTSAASPNIDD455 pKa = 3.6NFSGAHH461 pKa = 5.96SGLFSPNGIAVDD473 pKa = 3.77GAGNVWVTNYY483 pKa = 9.26YY484 pKa = 10.39QNPAGSLTEE493 pKa = 3.97VAGADD498 pKa = 3.31ASVPGAFLSPPDD510 pKa = 3.9AGFGSDD516 pKa = 5.14ASMLDD521 pKa = 3.48PYY523 pKa = 10.87GIAIDD528 pKa = 3.68ASGNLWVSNTGNLSVTQFVGAAVPVKK554 pKa = 9.32TPLVGPPQLPP564 pKa = 3.26
MM1 pKa = 7.09QLYY4 pKa = 10.37AAGTSGNGSAPQALLSSAVTTDD26 pKa = 2.91ATGSFTVPSGYY37 pKa = 8.65TCPSATAITYY47 pKa = 10.03LLAEE51 pKa = 5.28GGQAAGASSANADD64 pKa = 3.92LWLMAPIGACGDD76 pKa = 3.29ITASTTAVLNEE87 pKa = 3.97VTTVASAWALSQFLAPEE104 pKa = 4.62GNLGATSTNALGLANAVIVANNLVNIATGTSPGAAVPANVTVSTAKK150 pKa = 10.84LNTLADD156 pKa = 3.93ALSSCMAPAGEE167 pKa = 4.27TACAALLSAATTSGTTPGNTLDD189 pKa = 3.64AALNIARR196 pKa = 11.84HH197 pKa = 5.47PANNVSSIYY206 pKa = 10.46TLAQANAIFPSLTAAPPDD224 pKa = 3.22WLLYY228 pKa = 9.3RR229 pKa = 11.84TITGGGLEE237 pKa = 4.01QTPRR241 pKa = 11.84AIAIDD246 pKa = 3.48SAGNVWVATDD256 pKa = 3.41YY257 pKa = 11.55GAVAKK262 pKa = 10.41FSTNGTPVAPEE273 pKa = 4.28GFTGTGVNEE282 pKa = 4.28SRR284 pKa = 11.84SMALDD289 pKa = 3.06VHH291 pKa = 6.57DD292 pKa = 5.01NVWIANRR299 pKa = 11.84EE300 pKa = 4.06TTQNSGLGDD309 pKa = 4.09VIEE312 pKa = 4.85LSPTGQLLSGTGGFTDD328 pKa = 4.28GGINFPQGIATDD340 pKa = 4.03PNGNVWVANANLSTVTLLNDD360 pKa = 3.36SGTVLAASIGAGGGLIAVPDD380 pKa = 5.39AIAVDD385 pKa = 3.97ANDD388 pKa = 3.78NAWVGNTGAGSTITRR403 pKa = 11.84ISSDD407 pKa = 2.8GSQVTNLTCCDD418 pKa = 3.84HH419 pKa = 7.06PAGIAVDD426 pKa = 3.7QEE428 pKa = 4.79DD429 pKa = 4.45NVWTANFLDD438 pKa = 5.05DD439 pKa = 5.23SITMITNTSAASPNIDD455 pKa = 3.6NFSGAHH461 pKa = 5.96SGLFSPNGIAVDD473 pKa = 3.77GAGNVWVTNYY483 pKa = 9.26YY484 pKa = 10.39QNPAGSLTEE493 pKa = 3.97VAGADD498 pKa = 3.31ASVPGAFLSPPDD510 pKa = 3.9AGFGSDD516 pKa = 5.14ASMLDD521 pKa = 3.48PYY523 pKa = 10.87GIAIDD528 pKa = 3.68ASGNLWVSNTGNLSVTQFVGAAVPVKK554 pKa = 9.32TPLVGPPQLPP564 pKa = 3.26
Molecular weight: 56.01 kDa
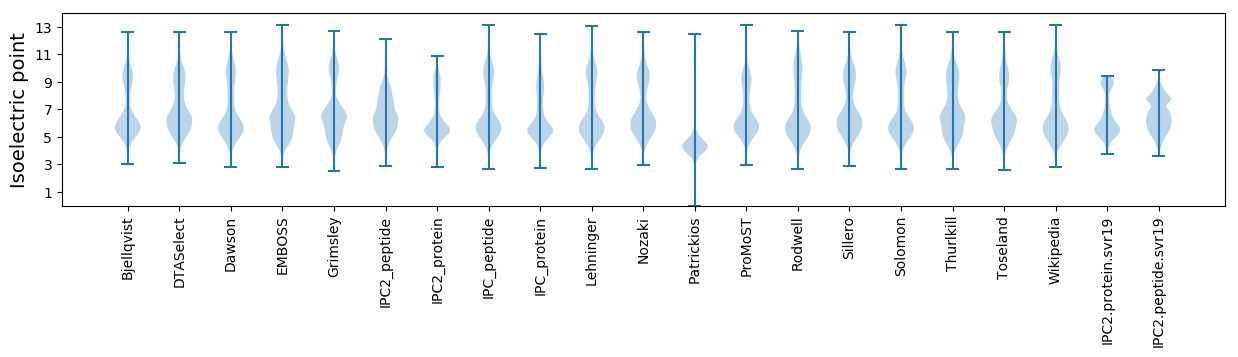
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8NZS6|G8NZS6_GRAMM Uncharacterized protein OS=Granulicella mallensis (strain ATCC BAA-1857 / DSM 23137 / MP5ACTX8) OX=682795 GN=AciX8_0195 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 9.81VHH17 pKa = 5.54GFLVRR22 pKa = 11.84MATKK26 pKa = 10.43AGQAVLNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 9.81VHH17 pKa = 5.54GFLVRR22 pKa = 11.84MATKK26 pKa = 10.43AGQAVLNRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
Molecular weight: 5.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1807496 |
30 |
5443 |
376.2 |
41.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.611 ± 0.038 | 0.85 ± 0.011 |
5.034 ± 0.018 | 5.273 ± 0.038 |
4.013 ± 0.026 | 8.116 ± 0.039 |
2.329 ± 0.017 | 4.999 ± 0.024 |
3.598 ± 0.029 | 10.033 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.016 | 3.641 ± 0.039 |
5.418 ± 0.026 | 3.923 ± 0.021 |
5.706 ± 0.036 | 6.728 ± 0.032 |
6.358 ± 0.037 | 7.046 ± 0.028 |
1.385 ± 0.015 | 2.826 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
