
Alloactinosynnema sp. L-07
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Alloactinosynnema; unclassified Alloactinosynnema
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
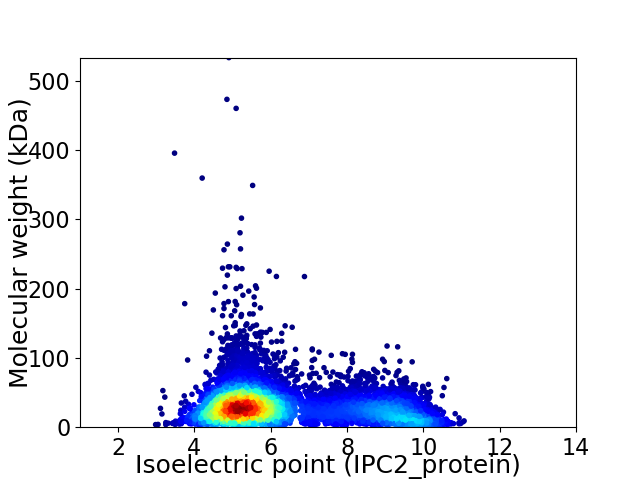
Virtual 2D-PAGE plot for 7267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H5CJT3|A0A0H5CJT3_9PSEU Uncharacterized protein OS=Alloactinosynnema sp. L-07 OX=1653480 PE=4 SV=1
MM1 pKa = 7.36SAATVWRR8 pKa = 11.84ASALAALVVVPSLVLTGTAQAQDD31 pKa = 3.36GTVQVISGEE40 pKa = 4.15LTIKK44 pKa = 10.63AAGRR48 pKa = 11.84QVNKK52 pKa = 10.24VLVSHH57 pKa = 6.48VGDD60 pKa = 3.6MVVVSDD66 pKa = 5.02VVPLSAGPGCRR77 pKa = 11.84KK78 pKa = 9.86YY79 pKa = 10.86EE80 pKa = 4.06RR81 pKa = 11.84TEE83 pKa = 3.92VRR85 pKa = 11.84CPATGVTSLAIDD97 pKa = 4.51LADD100 pKa = 4.4LDD102 pKa = 5.2DD103 pKa = 4.06RR104 pKa = 11.84AAYY107 pKa = 8.67KK108 pKa = 10.98GPMPVYY114 pKa = 10.64ADD116 pKa = 3.93GGAGDD121 pKa = 5.49DD122 pKa = 4.17IVTGGYY128 pKa = 10.5GGDD131 pKa = 3.46KK132 pKa = 10.69LLGGEE137 pKa = 4.24GVDD140 pKa = 3.98YY141 pKa = 11.42VSGGPGNDD149 pKa = 3.4TLIGGPGADD158 pKa = 2.79HH159 pKa = 7.16VYY161 pKa = 10.9GGTHH165 pKa = 6.64NDD167 pKa = 2.95VLYY170 pKa = 11.22GDD172 pKa = 5.18LPYY175 pKa = 10.13ATPLCDD181 pKa = 5.07SGLPSCGDD189 pKa = 3.33FLYY192 pKa = 11.04GGDD195 pKa = 4.36GADD198 pKa = 3.92KK199 pKa = 10.46IYY201 pKa = 10.78GGEE204 pKa = 4.04WADD207 pKa = 3.71TLAGEE212 pKa = 4.99GGNDD216 pKa = 3.78LLRR219 pKa = 11.84DD220 pKa = 4.22FSGGATFRR228 pKa = 11.84GGPGDD233 pKa = 3.59DD234 pKa = 3.53TMIGASPSMRR244 pKa = 11.84DD245 pKa = 3.16TVDD248 pKa = 3.44YY249 pKa = 10.93SDD251 pKa = 4.67HH252 pKa = 5.85LTDD255 pKa = 4.07VKK257 pKa = 11.33VDD259 pKa = 3.69LTAGTGGDD267 pKa = 3.8VQAGEE272 pKa = 4.81HH273 pKa = 6.03DD274 pKa = 4.94TITSIQDD281 pKa = 3.28VNGGAGNDD289 pKa = 3.67SLKK292 pKa = 11.19GNSGDD297 pKa = 4.53NNLTGNGGNDD307 pKa = 3.35SADD310 pKa = 3.6GGDD313 pKa = 3.31SVLYY317 pKa = 10.79NDD319 pKa = 3.71ICNVEE324 pKa = 4.29SPSYY328 pKa = 10.64CC329 pKa = 3.71
MM1 pKa = 7.36SAATVWRR8 pKa = 11.84ASALAALVVVPSLVLTGTAQAQDD31 pKa = 3.36GTVQVISGEE40 pKa = 4.15LTIKK44 pKa = 10.63AAGRR48 pKa = 11.84QVNKK52 pKa = 10.24VLVSHH57 pKa = 6.48VGDD60 pKa = 3.6MVVVSDD66 pKa = 5.02VVPLSAGPGCRR77 pKa = 11.84KK78 pKa = 9.86YY79 pKa = 10.86EE80 pKa = 4.06RR81 pKa = 11.84TEE83 pKa = 3.92VRR85 pKa = 11.84CPATGVTSLAIDD97 pKa = 4.51LADD100 pKa = 4.4LDD102 pKa = 5.2DD103 pKa = 4.06RR104 pKa = 11.84AAYY107 pKa = 8.67KK108 pKa = 10.98GPMPVYY114 pKa = 10.64ADD116 pKa = 3.93GGAGDD121 pKa = 5.49DD122 pKa = 4.17IVTGGYY128 pKa = 10.5GGDD131 pKa = 3.46KK132 pKa = 10.69LLGGEE137 pKa = 4.24GVDD140 pKa = 3.98YY141 pKa = 11.42VSGGPGNDD149 pKa = 3.4TLIGGPGADD158 pKa = 2.79HH159 pKa = 7.16VYY161 pKa = 10.9GGTHH165 pKa = 6.64NDD167 pKa = 2.95VLYY170 pKa = 11.22GDD172 pKa = 5.18LPYY175 pKa = 10.13ATPLCDD181 pKa = 5.07SGLPSCGDD189 pKa = 3.33FLYY192 pKa = 11.04GGDD195 pKa = 4.36GADD198 pKa = 3.92KK199 pKa = 10.46IYY201 pKa = 10.78GGEE204 pKa = 4.04WADD207 pKa = 3.71TLAGEE212 pKa = 4.99GGNDD216 pKa = 3.78LLRR219 pKa = 11.84DD220 pKa = 4.22FSGGATFRR228 pKa = 11.84GGPGDD233 pKa = 3.59DD234 pKa = 3.53TMIGASPSMRR244 pKa = 11.84DD245 pKa = 3.16TVDD248 pKa = 3.44YY249 pKa = 10.93SDD251 pKa = 4.67HH252 pKa = 5.85LTDD255 pKa = 4.07VKK257 pKa = 11.33VDD259 pKa = 3.69LTAGTGGDD267 pKa = 3.8VQAGEE272 pKa = 4.81HH273 pKa = 6.03DD274 pKa = 4.94TITSIQDD281 pKa = 3.28VNGGAGNDD289 pKa = 3.67SLKK292 pKa = 11.19GNSGDD297 pKa = 4.53NNLTGNGGNDD307 pKa = 3.35SADD310 pKa = 3.6GGDD313 pKa = 3.31SVLYY317 pKa = 10.79NDD319 pKa = 3.71ICNVEE324 pKa = 4.29SPSYY328 pKa = 10.64CC329 pKa = 3.71
Molecular weight: 33.14 kDa
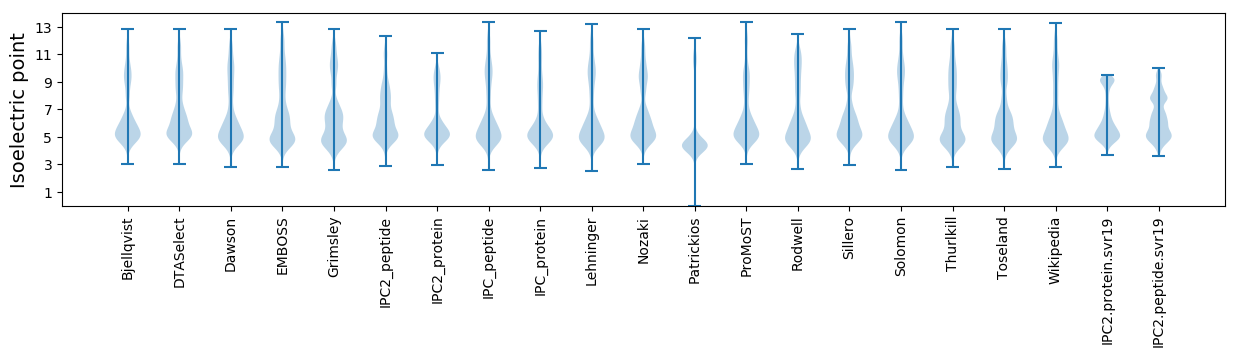
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H5CDQ8|A0A0H5CDQ8_9PSEU Uncharacterized protein OS=Alloactinosynnema sp. L-07 OX=1653480 PE=4 SV=1
MM1 pKa = 6.94GQGRR5 pKa = 11.84SFRR8 pKa = 11.84APLLAGRR15 pKa = 11.84GPLARR20 pKa = 11.84IHH22 pKa = 6.27PLVVFLLVAGLFAAGVLVRR41 pKa = 11.84GVLGAALLGVLAAGVAVLLAATWRR65 pKa = 11.84VQTASQRR72 pKa = 11.84VGRR75 pKa = 11.84VLVLGLLVAIAVSVLL90 pKa = 3.26
MM1 pKa = 6.94GQGRR5 pKa = 11.84SFRR8 pKa = 11.84APLLAGRR15 pKa = 11.84GPLARR20 pKa = 11.84IHH22 pKa = 6.27PLVVFLLVAGLFAAGVLVRR41 pKa = 11.84GVLGAALLGVLAAGVAVLLAATWRR65 pKa = 11.84VQTASQRR72 pKa = 11.84VGRR75 pKa = 11.84VLVLGLLVAIAVSVLL90 pKa = 3.26
Molecular weight: 9.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2223605 |
37 |
5032 |
306.0 |
32.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.339 ± 0.045 | 0.816 ± 0.01 |
6.423 ± 0.022 | 5.174 ± 0.028 |
2.821 ± 0.018 | 9.008 ± 0.029 |
2.259 ± 0.015 | 3.697 ± 0.021 |
2.194 ± 0.022 | 10.091 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.806 ± 0.013 | 2.0 ± 0.022 |
5.905 ± 0.026 | 2.723 ± 0.017 |
7.769 ± 0.035 | 5.14 ± 0.024 |
6.435 ± 0.038 | 8.931 ± 0.031 |
1.527 ± 0.012 | 1.943 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
