
Wuhan sharpbelly bornavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Bornaviridae; Cultervirus; Sharpbelly cultervirus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
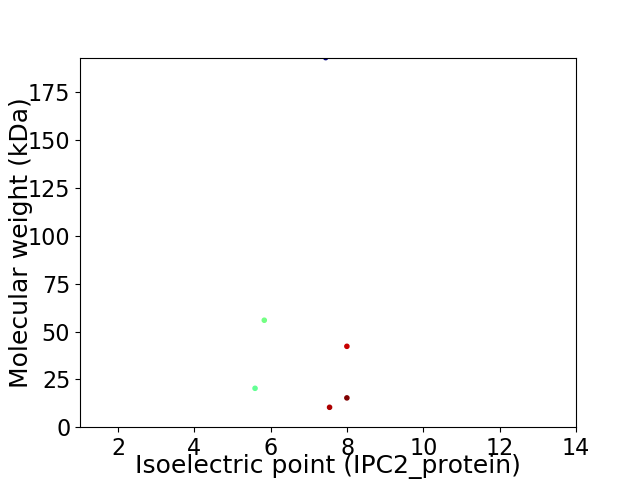
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GNX3|A0A2P1GNX3_9MONO Glycoprotein OS=Wuhan sharpbelly bornavirus OX=2116489 PE=4 SV=1
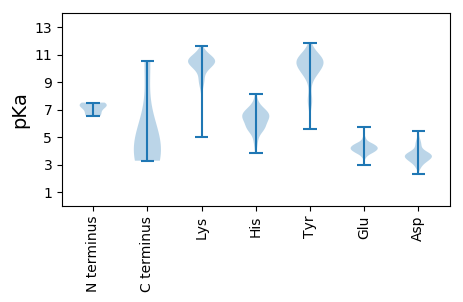
MM1 pKa = 7.28PQAPLLLLLLLPTITPFKK19 pKa = 10.11TLRR22 pKa = 11.84CDD24 pKa = 3.45TSSSPSAIKK33 pKa = 10.07YY34 pKa = 7.83VPPKK38 pKa = 10.53VDD40 pKa = 4.93LGLQNATCTVKK51 pKa = 10.41AKK53 pKa = 10.55KK54 pKa = 10.24RR55 pKa = 11.84ITEE58 pKa = 4.19GMQVEE63 pKa = 3.99IHH65 pKa = 6.3ACYY68 pKa = 10.46KK69 pKa = 8.61ITTYY73 pKa = 10.43TSWGVFQDD81 pKa = 3.69CDD83 pKa = 3.47AKK85 pKa = 11.29EE86 pKa = 4.37EE87 pKa = 4.14ITRR90 pKa = 11.84QYY92 pKa = 9.48PGCIVGDD99 pKa = 4.65EE100 pKa = 4.51IPSDD104 pKa = 3.9PCTAWWWRR112 pKa = 11.84GNGPDD117 pKa = 2.91IWKK120 pKa = 9.61LASQRR125 pKa = 11.84YY126 pKa = 5.61KK127 pKa = 10.84TEE129 pKa = 3.6VCICLPSVSATVAVSNHH146 pKa = 6.39PPPIYY151 pKa = 10.39CSFDD155 pKa = 3.52DD156 pKa = 5.46CSTCTVRR163 pKa = 11.84DD164 pKa = 3.82LAAGLCTLRR173 pKa = 11.84SMKK176 pKa = 10.22DD177 pKa = 2.79IKK179 pKa = 10.58FVPKK183 pKa = 10.26VLEE186 pKa = 4.33AEE188 pKa = 4.18EE189 pKa = 4.43QEE191 pKa = 4.24TDD193 pKa = 3.16EE194 pKa = 4.77SGVFHH199 pKa = 6.62FHH201 pKa = 5.83TRR203 pKa = 11.84HH204 pKa = 5.67IEE206 pKa = 3.68LPTFHH211 pKa = 7.27SSYY214 pKa = 10.98PLTADD219 pKa = 4.89LKK221 pKa = 11.22YY222 pKa = 10.76SDD224 pKa = 4.91ANIKK228 pKa = 9.91IEE230 pKa = 4.3CKK232 pKa = 9.79LVQRR236 pKa = 11.84PRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.5RR242 pKa = 11.84DD243 pKa = 3.44DD244 pKa = 3.13VQQYY248 pKa = 9.49VFDD251 pKa = 4.33ALAPHH256 pKa = 6.78LAEE259 pKa = 4.81SKK261 pKa = 7.36MTSWTISTLQDD272 pKa = 3.62TILSQPLIDD281 pKa = 3.53YY282 pKa = 7.32TKK284 pKa = 10.32YY285 pKa = 10.13VQAILKK291 pKa = 10.35RR292 pKa = 11.84NDD294 pKa = 2.86IVGTVNSGLVLYY306 pKa = 7.88WSCDD310 pKa = 3.32QVNADD315 pKa = 4.38FLPWTNDD322 pKa = 2.6TFYY325 pKa = 10.99PPVTVNGSLQYY336 pKa = 9.66LHH338 pKa = 7.92PYY340 pKa = 8.82TGILFSSSPPAPHH353 pKa = 6.75GLYY356 pKa = 10.67SLIYY360 pKa = 10.22VDD362 pKa = 3.57TKK364 pKa = 10.96HH365 pKa = 6.36YY366 pKa = 9.9LGSVGSGTTPHH377 pKa = 7.05TISTTHH383 pKa = 5.77VLSSAYY389 pKa = 10.19EE390 pKa = 4.08NVEE393 pKa = 3.92LDD395 pKa = 3.17ALQDD399 pKa = 3.62FRR401 pKa = 11.84LGSHH405 pKa = 6.72YY406 pKa = 10.36IDD408 pKa = 3.94PTSVAIGPLQDD419 pKa = 3.86PLAAAHH425 pKa = 6.32EE426 pKa = 4.51SSKK429 pKa = 11.01RR430 pKa = 11.84LLDD433 pKa = 3.67AAALHH438 pKa = 6.14SVGLGYY444 pKa = 10.61VYY446 pKa = 10.4QYY448 pKa = 11.87DD449 pKa = 3.84PMYY452 pKa = 10.41LIKK455 pKa = 10.78DD456 pKa = 3.87LLVKK460 pKa = 10.36VATIGGVMYY469 pKa = 9.82FCYY472 pKa = 10.58CMFICFKK479 pKa = 10.36YY480 pKa = 10.21LYY482 pKa = 10.18HH483 pKa = 6.83FIVWGQLILKK493 pKa = 9.14PPALIPP499 pKa = 4.1
MM1 pKa = 7.28PQAPLLLLLLLPTITPFKK19 pKa = 10.11TLRR22 pKa = 11.84CDD24 pKa = 3.45TSSSPSAIKK33 pKa = 10.07YY34 pKa = 7.83VPPKK38 pKa = 10.53VDD40 pKa = 4.93LGLQNATCTVKK51 pKa = 10.41AKK53 pKa = 10.55KK54 pKa = 10.24RR55 pKa = 11.84ITEE58 pKa = 4.19GMQVEE63 pKa = 3.99IHH65 pKa = 6.3ACYY68 pKa = 10.46KK69 pKa = 8.61ITTYY73 pKa = 10.43TSWGVFQDD81 pKa = 3.69CDD83 pKa = 3.47AKK85 pKa = 11.29EE86 pKa = 4.37EE87 pKa = 4.14ITRR90 pKa = 11.84QYY92 pKa = 9.48PGCIVGDD99 pKa = 4.65EE100 pKa = 4.51IPSDD104 pKa = 3.9PCTAWWWRR112 pKa = 11.84GNGPDD117 pKa = 2.91IWKK120 pKa = 9.61LASQRR125 pKa = 11.84YY126 pKa = 5.61KK127 pKa = 10.84TEE129 pKa = 3.6VCICLPSVSATVAVSNHH146 pKa = 6.39PPPIYY151 pKa = 10.39CSFDD155 pKa = 3.52DD156 pKa = 5.46CSTCTVRR163 pKa = 11.84DD164 pKa = 3.82LAAGLCTLRR173 pKa = 11.84SMKK176 pKa = 10.22DD177 pKa = 2.79IKK179 pKa = 10.58FVPKK183 pKa = 10.26VLEE186 pKa = 4.33AEE188 pKa = 4.18EE189 pKa = 4.43QEE191 pKa = 4.24TDD193 pKa = 3.16EE194 pKa = 4.77SGVFHH199 pKa = 6.62FHH201 pKa = 5.83TRR203 pKa = 11.84HH204 pKa = 5.67IEE206 pKa = 3.68LPTFHH211 pKa = 7.27SSYY214 pKa = 10.98PLTADD219 pKa = 4.89LKK221 pKa = 11.22YY222 pKa = 10.76SDD224 pKa = 4.91ANIKK228 pKa = 9.91IEE230 pKa = 4.3CKK232 pKa = 9.79LVQRR236 pKa = 11.84PRR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.5RR242 pKa = 11.84DD243 pKa = 3.44DD244 pKa = 3.13VQQYY248 pKa = 9.49VFDD251 pKa = 4.33ALAPHH256 pKa = 6.78LAEE259 pKa = 4.81SKK261 pKa = 7.36MTSWTISTLQDD272 pKa = 3.62TILSQPLIDD281 pKa = 3.53YY282 pKa = 7.32TKK284 pKa = 10.32YY285 pKa = 10.13VQAILKK291 pKa = 10.35RR292 pKa = 11.84NDD294 pKa = 2.86IVGTVNSGLVLYY306 pKa = 7.88WSCDD310 pKa = 3.32QVNADD315 pKa = 4.38FLPWTNDD322 pKa = 2.6TFYY325 pKa = 10.99PPVTVNGSLQYY336 pKa = 9.66LHH338 pKa = 7.92PYY340 pKa = 8.82TGILFSSSPPAPHH353 pKa = 6.75GLYY356 pKa = 10.67SLIYY360 pKa = 10.22VDD362 pKa = 3.57TKK364 pKa = 10.96HH365 pKa = 6.36YY366 pKa = 9.9LGSVGSGTTPHH377 pKa = 7.05TISTTHH383 pKa = 5.77VLSSAYY389 pKa = 10.19EE390 pKa = 4.08NVEE393 pKa = 3.92LDD395 pKa = 3.17ALQDD399 pKa = 3.62FRR401 pKa = 11.84LGSHH405 pKa = 6.72YY406 pKa = 10.36IDD408 pKa = 3.94PTSVAIGPLQDD419 pKa = 3.86PLAAAHH425 pKa = 6.32EE426 pKa = 4.51SSKK429 pKa = 11.01RR430 pKa = 11.84LLDD433 pKa = 3.67AAALHH438 pKa = 6.14SVGLGYY444 pKa = 10.61VYY446 pKa = 10.4QYY448 pKa = 11.87DD449 pKa = 3.84PMYY452 pKa = 10.41LIKK455 pKa = 10.78DD456 pKa = 3.87LLVKK460 pKa = 10.36VATIGGVMYY469 pKa = 9.82FCYY472 pKa = 10.58CMFICFKK479 pKa = 10.36YY480 pKa = 10.21LYY482 pKa = 10.18HH483 pKa = 6.83FIVWGQLILKK493 pKa = 9.14PPALIPP499 pKa = 4.1
Molecular weight: 55.91 kDa
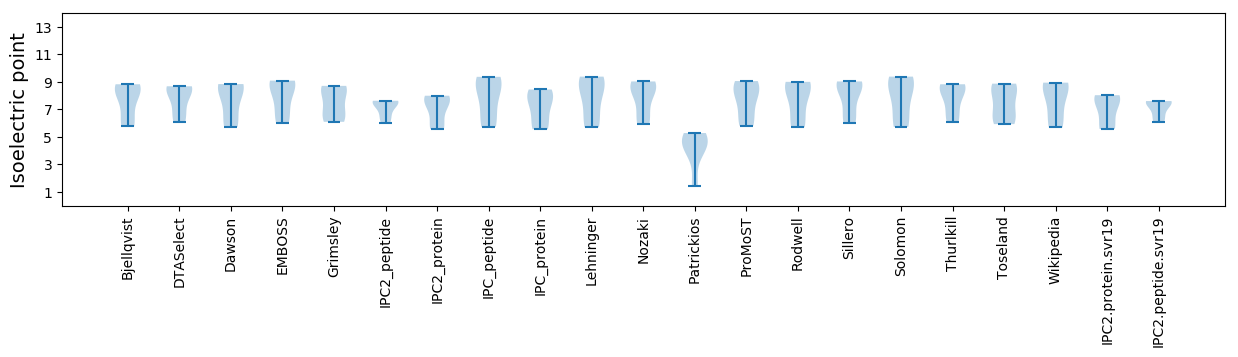
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GNX3|A0A2P1GNX3_9MONO Glycoprotein OS=Wuhan sharpbelly bornavirus OX=2116489 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 9.78MAGKK6 pKa = 10.27YY7 pKa = 7.25PTVDD11 pKa = 3.38PEE13 pKa = 3.87KK14 pKa = 10.96SIVQGYY20 pKa = 10.03HH21 pKa = 5.83SLLLHH26 pKa = 6.04VEE28 pKa = 4.32FRR30 pKa = 11.84DD31 pKa = 3.43SKK33 pKa = 11.22QFMKK37 pKa = 10.71FALTSTTTTFHH48 pKa = 6.17VPRR51 pKa = 11.84TTAVLTIDD59 pKa = 4.64FEE61 pKa = 4.46PANQEE66 pKa = 4.26DD67 pKa = 3.41VWMYY71 pKa = 9.79YY72 pKa = 7.9EE73 pKa = 4.52WEE75 pKa = 4.08PTTYY79 pKa = 10.94SSLKK83 pKa = 10.6LILPKK88 pKa = 10.28PGQFPISGKK97 pKa = 9.68IRR99 pKa = 11.84LSGEE103 pKa = 4.1AGQHH107 pKa = 6.36ALRR110 pKa = 11.84CDD112 pKa = 4.03FNVADD117 pKa = 3.99YY118 pKa = 10.78RR119 pKa = 11.84IISVPPGPLMHH130 pKa = 6.56RR131 pKa = 11.84VVRR134 pKa = 4.25
MM1 pKa = 7.4KK2 pKa = 9.78MAGKK6 pKa = 10.27YY7 pKa = 7.25PTVDD11 pKa = 3.38PEE13 pKa = 3.87KK14 pKa = 10.96SIVQGYY20 pKa = 10.03HH21 pKa = 5.83SLLLHH26 pKa = 6.04VEE28 pKa = 4.32FRR30 pKa = 11.84DD31 pKa = 3.43SKK33 pKa = 11.22QFMKK37 pKa = 10.71FALTSTTTTFHH48 pKa = 6.17VPRR51 pKa = 11.84TTAVLTIDD59 pKa = 4.64FEE61 pKa = 4.46PANQEE66 pKa = 4.26DD67 pKa = 3.41VWMYY71 pKa = 9.79YY72 pKa = 7.9EE73 pKa = 4.52WEE75 pKa = 4.08PTTYY79 pKa = 10.94SSLKK83 pKa = 10.6LILPKK88 pKa = 10.28PGQFPISGKK97 pKa = 9.68IRR99 pKa = 11.84LSGEE103 pKa = 4.1AGQHH107 pKa = 6.36ALRR110 pKa = 11.84CDD112 pKa = 4.03FNVADD117 pKa = 3.99YY118 pKa = 10.78RR119 pKa = 11.84IISVPPGPLMHH130 pKa = 6.56RR131 pKa = 11.84VVRR134 pKa = 4.25
Molecular weight: 15.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3030 |
98 |
1729 |
505.0 |
56.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.469 ± 0.791 | 2.145 ± 0.528 |
4.752 ± 0.401 | 5.38 ± 0.774 |
3.234 ± 0.49 | 5.908 ± 0.595 |
2.904 ± 0.171 | 5.578 ± 0.23 |
5.314 ± 0.615 | 10.561 ± 1.431 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.32 | 2.739 ± 0.239 |
5.71 ± 0.744 | 4.224 ± 0.269 |
5.182 ± 0.498 | 8.614 ± 0.349 |
7.195 ± 0.341 | 7.063 ± 0.235 |
1.089 ± 0.237 | 3.531 ± 0.573 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
