
Achromobacter phage phiAxp-3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Rothmandenesvirinae; Jwalphavirus; Achromobacter virus Axp3
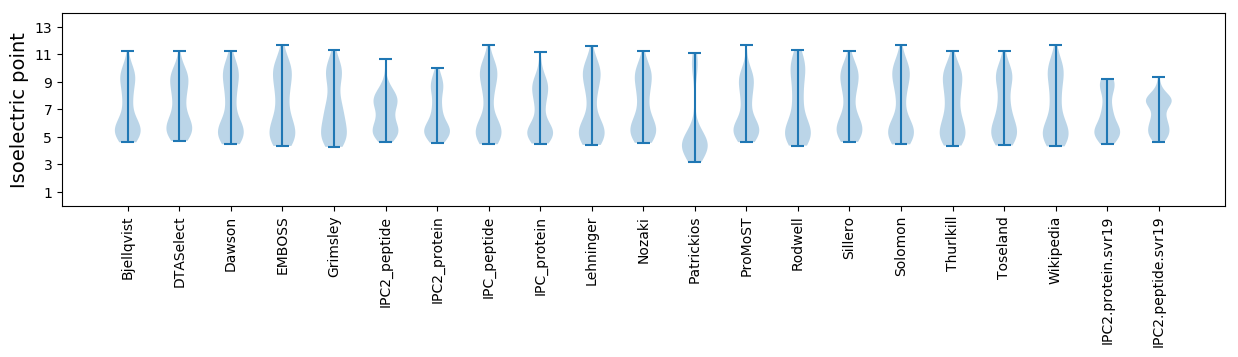
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0K2FHI7|A0A0K2FHI7_9CAUD Uncharacterized protein OS=Achromobacter phage phiAxp-3 OX=1664247 GN=ADP65_00073 PE=4 SV=1
MM1 pKa = 7.25GVAIDD6 pKa = 4.54MATQDD11 pKa = 3.98AVVAMPEE18 pKa = 3.82AALEE22 pKa = 4.06EE23 pKa = 4.15QQVLIRR29 pKa = 11.84QMQAEE34 pKa = 5.06LVAEE38 pKa = 4.28MPGLILGAHH47 pKa = 5.48TVQYY51 pKa = 10.7GPNDD55 pKa = 3.44YY56 pKa = 11.02AFEE59 pKa = 4.63PGIGQIHH66 pKa = 6.18YY67 pKa = 10.63LPATSQRR74 pKa = 11.84ICKK77 pKa = 10.12DD78 pKa = 3.05LAQSYY83 pKa = 9.65LALIGDD89 pKa = 4.35YY90 pKa = 10.82PLVSPIKK97 pKa = 8.47PTRR100 pKa = 11.84DD101 pKa = 2.9SYY103 pKa = 11.9EE104 pKa = 3.76FVGRR108 pKa = 11.84LGIDD112 pKa = 3.22PVNEE116 pKa = 4.06PEE118 pKa = 4.18PVNWPTMEE126 pKa = 3.91YY127 pKa = 10.09YY128 pKa = 10.45RR129 pKa = 11.84RR130 pKa = 11.84FLRR133 pKa = 11.84LLHH136 pKa = 5.83EE137 pKa = 4.82RR138 pKa = 11.84NFKK141 pKa = 10.11FVNSVAYY148 pKa = 9.71EE149 pKa = 3.76ILDD152 pKa = 3.38FFMPEE157 pKa = 3.28EE158 pKa = 3.95WKK160 pKa = 10.19QRR162 pKa = 11.84NWLGLPAQSGWYY174 pKa = 8.25PPSSFIQPANEE185 pKa = 4.0APLDD189 pKa = 4.09YY190 pKa = 10.1IAKK193 pKa = 9.63VQIQILKK200 pKa = 9.31AGQEE204 pKa = 4.04IGMVPYY210 pKa = 9.77FQIGEE215 pKa = 4.17PWWWDD220 pKa = 2.96GSYY223 pKa = 11.66NNGEE227 pKa = 4.42GKK229 pKa = 9.65NAPCLYY235 pKa = 10.19DD236 pKa = 3.57QRR238 pKa = 11.84TMDD241 pKa = 4.67LYY243 pKa = 11.12KK244 pKa = 10.42QEE246 pKa = 4.32TGNDD250 pKa = 3.01VPMPMIKK257 pKa = 10.3SIFDD261 pKa = 3.7PVADD265 pKa = 4.15NQWPYY270 pKa = 11.04IDD272 pKa = 3.58WLRR275 pKa = 11.84TKK277 pKa = 10.8LGDD280 pKa = 3.28STNYY284 pKa = 9.59VRR286 pKa = 11.84DD287 pKa = 3.55KK288 pKa = 11.34VKK290 pKa = 10.67AAFPNAQATLLFFSPQVMSPASDD313 pKa = 3.15LTFRR317 pKa = 11.84LNFPMDD323 pKa = 2.86EE324 pKa = 3.99WVYY327 pKa = 10.58PNYY330 pKa = 10.58EE331 pKa = 3.88FVQIEE336 pKa = 4.4DD337 pKa = 3.92YY338 pKa = 11.37DD339 pKa = 4.01WIIDD343 pKa = 3.74GRR345 pKa = 11.84LDD347 pKa = 4.43LVPQTFTAATEE358 pKa = 3.94LLKK361 pKa = 11.21YY362 pKa = 9.41PLEE365 pKa = 4.22VVHH368 pKa = 6.01YY369 pKa = 8.29FIGFTLLPEE378 pKa = 4.2DD379 pKa = 5.1AKK381 pKa = 11.07RR382 pKa = 11.84IWGNTDD388 pKa = 3.21KK389 pKa = 11.24AWALAQEE396 pKa = 4.39AGIKK400 pKa = 9.72IIYY403 pKa = 7.48PWSYY407 pKa = 7.66TQVMRR412 pKa = 11.84DD413 pKa = 3.08GVYY416 pKa = 9.15FARR419 pKa = 11.84PKK421 pKa = 10.56VCGCC425 pKa = 4.1
MM1 pKa = 7.25GVAIDD6 pKa = 4.54MATQDD11 pKa = 3.98AVVAMPEE18 pKa = 3.82AALEE22 pKa = 4.06EE23 pKa = 4.15QQVLIRR29 pKa = 11.84QMQAEE34 pKa = 5.06LVAEE38 pKa = 4.28MPGLILGAHH47 pKa = 5.48TVQYY51 pKa = 10.7GPNDD55 pKa = 3.44YY56 pKa = 11.02AFEE59 pKa = 4.63PGIGQIHH66 pKa = 6.18YY67 pKa = 10.63LPATSQRR74 pKa = 11.84ICKK77 pKa = 10.12DD78 pKa = 3.05LAQSYY83 pKa = 9.65LALIGDD89 pKa = 4.35YY90 pKa = 10.82PLVSPIKK97 pKa = 8.47PTRR100 pKa = 11.84DD101 pKa = 2.9SYY103 pKa = 11.9EE104 pKa = 3.76FVGRR108 pKa = 11.84LGIDD112 pKa = 3.22PVNEE116 pKa = 4.06PEE118 pKa = 4.18PVNWPTMEE126 pKa = 3.91YY127 pKa = 10.09YY128 pKa = 10.45RR129 pKa = 11.84RR130 pKa = 11.84FLRR133 pKa = 11.84LLHH136 pKa = 5.83EE137 pKa = 4.82RR138 pKa = 11.84NFKK141 pKa = 10.11FVNSVAYY148 pKa = 9.71EE149 pKa = 3.76ILDD152 pKa = 3.38FFMPEE157 pKa = 3.28EE158 pKa = 3.95WKK160 pKa = 10.19QRR162 pKa = 11.84NWLGLPAQSGWYY174 pKa = 8.25PPSSFIQPANEE185 pKa = 4.0APLDD189 pKa = 4.09YY190 pKa = 10.1IAKK193 pKa = 9.63VQIQILKK200 pKa = 9.31AGQEE204 pKa = 4.04IGMVPYY210 pKa = 9.77FQIGEE215 pKa = 4.17PWWWDD220 pKa = 2.96GSYY223 pKa = 11.66NNGEE227 pKa = 4.42GKK229 pKa = 9.65NAPCLYY235 pKa = 10.19DD236 pKa = 3.57QRR238 pKa = 11.84TMDD241 pKa = 4.67LYY243 pKa = 11.12KK244 pKa = 10.42QEE246 pKa = 4.32TGNDD250 pKa = 3.01VPMPMIKK257 pKa = 10.3SIFDD261 pKa = 3.7PVADD265 pKa = 4.15NQWPYY270 pKa = 11.04IDD272 pKa = 3.58WLRR275 pKa = 11.84TKK277 pKa = 10.8LGDD280 pKa = 3.28STNYY284 pKa = 9.59VRR286 pKa = 11.84DD287 pKa = 3.55KK288 pKa = 11.34VKK290 pKa = 10.67AAFPNAQATLLFFSPQVMSPASDD313 pKa = 3.15LTFRR317 pKa = 11.84LNFPMDD323 pKa = 2.86EE324 pKa = 3.99WVYY327 pKa = 10.58PNYY330 pKa = 10.58EE331 pKa = 3.88FVQIEE336 pKa = 4.4DD337 pKa = 3.92YY338 pKa = 11.37DD339 pKa = 4.01WIIDD343 pKa = 3.74GRR345 pKa = 11.84LDD347 pKa = 4.43LVPQTFTAATEE358 pKa = 3.94LLKK361 pKa = 11.21YY362 pKa = 9.41PLEE365 pKa = 4.22VVHH368 pKa = 6.01YY369 pKa = 8.29FIGFTLLPEE378 pKa = 4.2DD379 pKa = 5.1AKK381 pKa = 11.07RR382 pKa = 11.84IWGNTDD388 pKa = 3.21KK389 pKa = 11.24AWALAQEE396 pKa = 4.39AGIKK400 pKa = 9.72IIYY403 pKa = 7.48PWSYY407 pKa = 7.66TQVMRR412 pKa = 11.84DD413 pKa = 3.08GVYY416 pKa = 9.15FARR419 pKa = 11.84PKK421 pKa = 10.56VCGCC425 pKa = 4.1
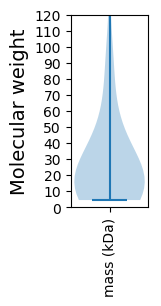
Molecular weight: 48.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K2FI96|A0A0K2FI96_9CAUD Uncharacterized protein OS=Achromobacter phage phiAxp-3 OX=1664247 GN=ADP65_00026 PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 5.41KK3 pKa = 10.21NRR5 pKa = 11.84RR6 pKa = 11.84NAIIEE11 pKa = 4.41KK12 pKa = 10.09IEE14 pKa = 3.92ARR16 pKa = 11.84CDD18 pKa = 3.26IVDD21 pKa = 3.37MGFVIGMKK29 pKa = 10.16PSPCHH34 pKa = 6.1LWQGPTSGNGRR45 pKa = 11.84GGGYY49 pKa = 10.88GRR51 pKa = 11.84MSLNGHH57 pKa = 5.49TVAVHH62 pKa = 5.85LVVFTHH68 pKa = 5.85YY69 pKa = 10.42FGYY72 pKa = 10.25IPGNKK77 pKa = 8.97QVDD80 pKa = 3.82HH81 pKa = 6.98LCNQRR86 pKa = 11.84LCCNPQHH93 pKa = 6.7LEE95 pKa = 4.09LVTHH99 pKa = 6.71LTNQRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84AARR109 pKa = 11.84SKK111 pKa = 10.85SS112 pKa = 3.34
MM1 pKa = 7.53EE2 pKa = 5.41KK3 pKa = 10.21NRR5 pKa = 11.84RR6 pKa = 11.84NAIIEE11 pKa = 4.41KK12 pKa = 10.09IEE14 pKa = 3.92ARR16 pKa = 11.84CDD18 pKa = 3.26IVDD21 pKa = 3.37MGFVIGMKK29 pKa = 10.16PSPCHH34 pKa = 6.1LWQGPTSGNGRR45 pKa = 11.84GGGYY49 pKa = 10.88GRR51 pKa = 11.84MSLNGHH57 pKa = 5.49TVAVHH62 pKa = 5.85LVVFTHH68 pKa = 5.85YY69 pKa = 10.42FGYY72 pKa = 10.25IPGNKK77 pKa = 8.97QVDD80 pKa = 3.82HH81 pKa = 6.98LCNQRR86 pKa = 11.84LCCNPQHH93 pKa = 6.7LEE95 pKa = 4.09LVTHH99 pKa = 6.71LTNQRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84AARR109 pKa = 11.84SKK111 pKa = 10.85SS112 pKa = 3.34
Molecular weight: 12.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
22633 |
39 |
3428 |
282.9 |
31.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.517 ± 0.614 | 0.742 ± 0.131 |
5.828 ± 0.151 | 6.292 ± 0.184 |
3.636 ± 0.196 | 7.171 ± 0.236 |
1.94 ± 0.173 | 5.112 ± 0.214 |
6.027 ± 0.237 | 8.474 ± 0.255 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.358 ± 0.131 | 4.71 ± 0.165 |
4.617 ± 0.224 | 4.975 ± 0.216 |
4.918 ± 0.182 | 5.465 ± 0.21 |
5.912 ± 0.209 | 6.689 ± 0.209 |
1.458 ± 0.182 | 3.159 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
