
Roseobacter sp. AzwK-3b
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseobacter; unclassified Roseobacter
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
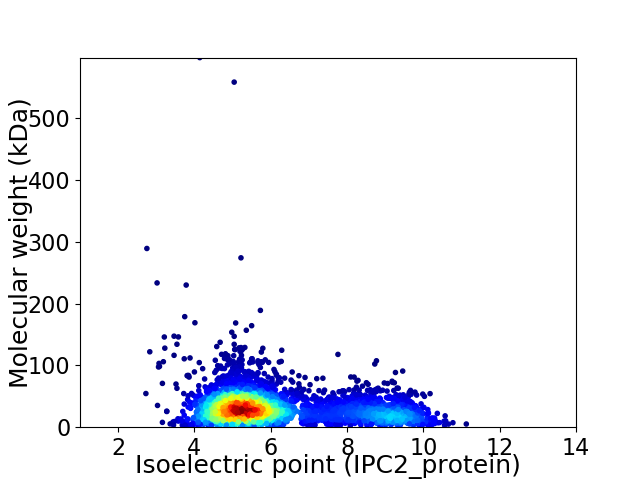
Virtual 2D-PAGE plot for 4072 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6FWK1|A6FWK1_9RHOB Uncharacterized protein OS=Roseobacter sp. AzwK-3b OX=351016 GN=RAZWK3B_11642 PE=4 SV=1
MM1 pKa = 7.17VGGAASAQDD10 pKa = 3.51WNLDD14 pKa = 2.84WGGYY18 pKa = 9.0YY19 pKa = 11.06NMDD22 pKa = 3.28VLYY25 pKa = 10.87NDD27 pKa = 3.65ASGNFLPAAVDD38 pKa = 3.37VDD40 pKa = 3.76GVNIHH45 pKa = 5.97QNGEE49 pKa = 3.75IHH51 pKa = 6.06FTPSVTLDD59 pKa = 2.81NGMTFGVVVEE69 pKa = 4.66MEE71 pKa = 4.03AANAGGGADD80 pKa = 5.17GIDD83 pKa = 3.71EE84 pKa = 4.75SYY86 pKa = 10.68MFISSDD92 pKa = 2.93SMGRR96 pKa = 11.84VEE98 pKa = 5.13VGSEE102 pKa = 3.45NSAGYY107 pKa = 9.25KK108 pKa = 10.17LSVGAPSVGFGINSPSPSAYY128 pKa = 10.13HH129 pKa = 5.37PTAFLFPDD137 pKa = 4.89ADD139 pKa = 3.92PADD142 pKa = 3.56ATTIGEE148 pKa = 4.55YY149 pKa = 9.71IDD151 pKa = 5.34ALDD154 pKa = 4.69GVDD157 pKa = 4.3NPLLGGAGFALGRR170 pKa = 11.84NAMGSSATEE179 pKa = 3.61VAGNNDD185 pKa = 3.6TARR188 pKa = 11.84ISYY191 pKa = 7.37YY192 pKa = 9.79TPSFNGLTLGVSYY205 pKa = 11.2APGNTNVANSGLVNRR220 pKa = 11.84NGLSDD225 pKa = 3.44IFDD228 pKa = 3.27IGANYY233 pKa = 9.66SQSFGNVDD241 pKa = 3.39MTLSARR247 pKa = 11.84WGTADD252 pKa = 3.57AATARR257 pKa = 11.84STAAGTTGPGLDD269 pKa = 3.98GVAGTADD276 pKa = 3.82DD277 pKa = 4.41VYY279 pKa = 11.43AAAVVQQDD287 pKa = 5.13DD288 pKa = 3.94PTTWAIGAQFATAGFTFGGSYY309 pKa = 10.98AEE311 pKa = 4.2NDD313 pKa = 3.76AGSVNADD320 pKa = 3.57GLKK323 pKa = 10.83DD324 pKa = 3.61GAGDD328 pKa = 3.64SSGWSLGVTYY338 pKa = 10.51DD339 pKa = 3.8VAGPWTIGFDD349 pKa = 3.72TYY351 pKa = 10.79QGEE354 pKa = 4.48MDD356 pKa = 4.04GSLDD360 pKa = 3.4FATGTRR366 pKa = 11.84TKK368 pKa = 10.7AEE370 pKa = 3.95YY371 pKa = 9.98VAYY374 pKa = 10.34QIGAEE379 pKa = 3.78RR380 pKa = 11.84SLGNGVAWRR389 pKa = 11.84IYY391 pKa = 10.19AIQSEE396 pKa = 4.8TSLKK400 pKa = 8.87TNRR403 pKa = 11.84IGGFANDD410 pKa = 3.41TGGNSNVSKK419 pKa = 11.07YY420 pKa = 10.21KK421 pKa = 10.89LEE423 pKa = 4.03STTIGTGIRR432 pKa = 11.84LTFF435 pKa = 4.02
MM1 pKa = 7.17VGGAASAQDD10 pKa = 3.51WNLDD14 pKa = 2.84WGGYY18 pKa = 9.0YY19 pKa = 11.06NMDD22 pKa = 3.28VLYY25 pKa = 10.87NDD27 pKa = 3.65ASGNFLPAAVDD38 pKa = 3.37VDD40 pKa = 3.76GVNIHH45 pKa = 5.97QNGEE49 pKa = 3.75IHH51 pKa = 6.06FTPSVTLDD59 pKa = 2.81NGMTFGVVVEE69 pKa = 4.66MEE71 pKa = 4.03AANAGGGADD80 pKa = 5.17GIDD83 pKa = 3.71EE84 pKa = 4.75SYY86 pKa = 10.68MFISSDD92 pKa = 2.93SMGRR96 pKa = 11.84VEE98 pKa = 5.13VGSEE102 pKa = 3.45NSAGYY107 pKa = 9.25KK108 pKa = 10.17LSVGAPSVGFGINSPSPSAYY128 pKa = 10.13HH129 pKa = 5.37PTAFLFPDD137 pKa = 4.89ADD139 pKa = 3.92PADD142 pKa = 3.56ATTIGEE148 pKa = 4.55YY149 pKa = 9.71IDD151 pKa = 5.34ALDD154 pKa = 4.69GVDD157 pKa = 4.3NPLLGGAGFALGRR170 pKa = 11.84NAMGSSATEE179 pKa = 3.61VAGNNDD185 pKa = 3.6TARR188 pKa = 11.84ISYY191 pKa = 7.37YY192 pKa = 9.79TPSFNGLTLGVSYY205 pKa = 11.2APGNTNVANSGLVNRR220 pKa = 11.84NGLSDD225 pKa = 3.44IFDD228 pKa = 3.27IGANYY233 pKa = 9.66SQSFGNVDD241 pKa = 3.39MTLSARR247 pKa = 11.84WGTADD252 pKa = 3.57AATARR257 pKa = 11.84STAAGTTGPGLDD269 pKa = 3.98GVAGTADD276 pKa = 3.82DD277 pKa = 4.41VYY279 pKa = 11.43AAAVVQQDD287 pKa = 5.13DD288 pKa = 3.94PTTWAIGAQFATAGFTFGGSYY309 pKa = 10.98AEE311 pKa = 4.2NDD313 pKa = 3.76AGSVNADD320 pKa = 3.57GLKK323 pKa = 10.83DD324 pKa = 3.61GAGDD328 pKa = 3.64SSGWSLGVTYY338 pKa = 10.51DD339 pKa = 3.8VAGPWTIGFDD349 pKa = 3.72TYY351 pKa = 10.79QGEE354 pKa = 4.48MDD356 pKa = 4.04GSLDD360 pKa = 3.4FATGTRR366 pKa = 11.84TKK368 pKa = 10.7AEE370 pKa = 3.95YY371 pKa = 9.98VAYY374 pKa = 10.34QIGAEE379 pKa = 3.78RR380 pKa = 11.84SLGNGVAWRR389 pKa = 11.84IYY391 pKa = 10.19AIQSEE396 pKa = 4.8TSLKK400 pKa = 8.87TNRR403 pKa = 11.84IGGFANDD410 pKa = 3.41TGGNSNVSKK419 pKa = 11.07YY420 pKa = 10.21KK421 pKa = 10.89LEE423 pKa = 4.03STTIGTGIRR432 pKa = 11.84LTFF435 pKa = 4.02
Molecular weight: 44.64 kDa
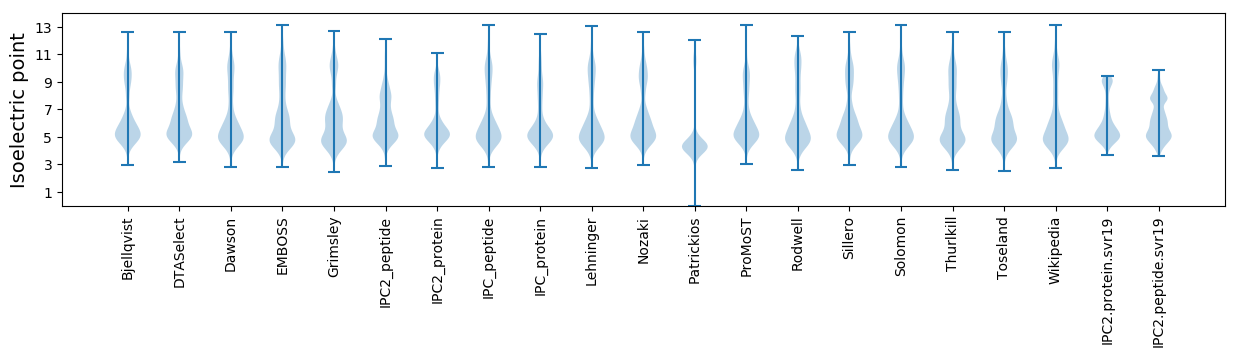
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6FS44|A6FS44_9RHOB Rieske 2Fe-2S domain protein putative OS=Roseobacter sp. AzwK-3b OX=351016 GN=RAZWK3B_16475 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1220550 |
17 |
5954 |
299.7 |
32.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.147 ± 0.055 | 0.925 ± 0.015 |
6.326 ± 0.041 | 5.864 ± 0.04 |
3.608 ± 0.027 | 8.66 ± 0.051 |
2.138 ± 0.027 | 5.222 ± 0.025 |
3.014 ± 0.033 | 10.015 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.844 ± 0.022 | 2.538 ± 0.025 |
5.022 ± 0.031 | 3.316 ± 0.024 |
6.951 ± 0.045 | 5.121 ± 0.033 |
5.528 ± 0.045 | 7.187 ± 0.034 |
1.382 ± 0.017 | 2.192 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
