
Changjiang tombus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
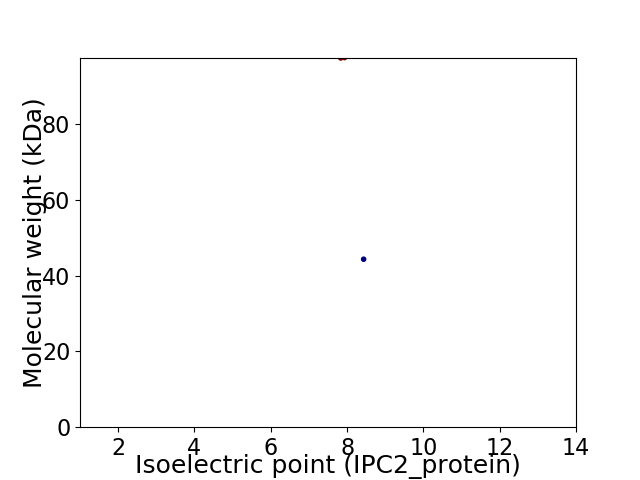
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
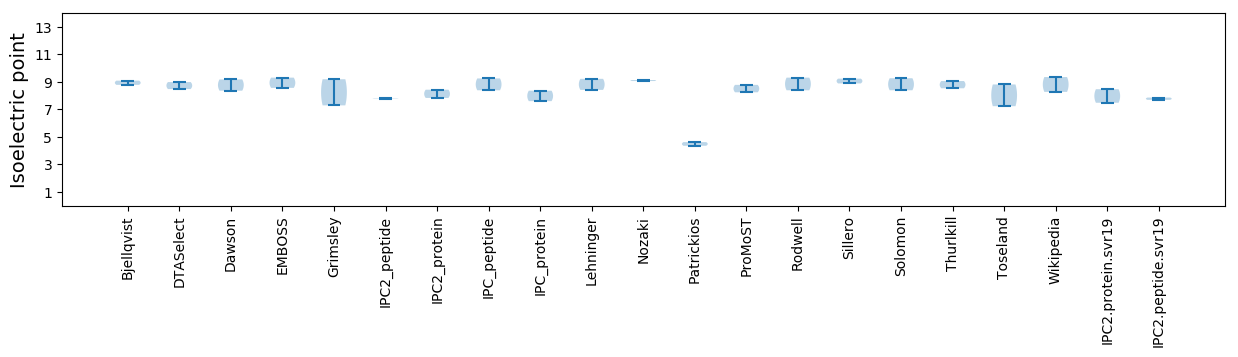
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFV5|A0A1L3KFV5_9VIRU Capsid protein OS=Changjiang tombus-like virus 9 OX=1922823 PE=3 SV=1
MM1 pKa = 7.64SFFEE5 pKa = 4.45LLRR8 pKa = 11.84AFLCCATADD17 pKa = 3.75SGEE20 pKa = 4.08VKK22 pKa = 10.0MDD24 pKa = 3.33VAKK27 pKa = 9.89TLLRR31 pKa = 11.84EE32 pKa = 4.33DD33 pKa = 4.77CLATDD38 pKa = 4.49NIEE41 pKa = 4.1FTHH44 pKa = 6.87DD45 pKa = 3.63PKK47 pKa = 10.59EE48 pKa = 3.95HH49 pKa = 6.21HH50 pKa = 5.52VTRR53 pKa = 11.84RR54 pKa = 11.84PAGVRR59 pKa = 11.84VTDD62 pKa = 3.49NRR64 pKa = 11.84EE65 pKa = 3.61ISPLEE70 pKa = 3.88PRR72 pKa = 11.84AGLPVKK78 pKa = 10.18KK79 pKa = 9.8CAYY82 pKa = 8.25RR83 pKa = 11.84AEE85 pKa = 4.1MDD87 pKa = 3.57VLRR90 pKa = 11.84KK91 pKa = 9.75LRR93 pKa = 11.84LAAEE97 pKa = 4.37DD98 pKa = 3.44AFGRR102 pKa = 11.84QQEE105 pKa = 4.41KK106 pKa = 10.24KK107 pKa = 10.17QPDD110 pKa = 4.03PVTQHH115 pKa = 7.02RR116 pKa = 11.84LEE118 pKa = 4.42CVLNKK123 pKa = 9.46ATLAAFKK130 pKa = 10.92VVGATHH136 pKa = 5.86QCEE139 pKa = 4.32SWSLLVAIGEE149 pKa = 4.31LQLDD153 pKa = 3.87MTHH156 pKa = 6.65TRR158 pKa = 11.84SEE160 pKa = 3.85WKK162 pKa = 10.05RR163 pKa = 11.84RR164 pKa = 11.84LLRR167 pKa = 11.84LGEE170 pKa = 4.16LCSIEE175 pKa = 5.15SGCEE179 pKa = 3.59WEE181 pKa = 5.15VEE183 pKa = 4.3GNPPGLASEE192 pKa = 4.13PHH194 pKa = 6.37PPSGEE199 pKa = 4.06VPVPTDD205 pKa = 2.99QPTGVHH211 pKa = 6.68RR212 pKa = 11.84LCHH215 pKa = 6.11DD216 pKa = 3.61VVEE219 pKa = 5.18VKK221 pKa = 10.19AHH223 pKa = 4.96RR224 pKa = 11.84RR225 pKa = 11.84IRR227 pKa = 11.84KK228 pKa = 9.11DD229 pKa = 3.03RR230 pKa = 11.84EE231 pKa = 3.85GQYY234 pKa = 10.09VGTVVRR240 pKa = 11.84EE241 pKa = 3.8LKK243 pKa = 10.81NRR245 pKa = 11.84FGCPKK250 pKa = 8.82PTEE253 pKa = 4.56DD254 pKa = 3.55NLLAVRR260 pKa = 11.84RR261 pKa = 11.84SAINIMEE268 pKa = 4.16KK269 pKa = 10.22HH270 pKa = 5.65GVRR273 pKa = 11.84PSHH276 pKa = 6.77VGEE279 pKa = 4.2VVDD282 pKa = 4.98LVVAGVFVPDD292 pKa = 3.53RR293 pKa = 11.84NEE295 pKa = 3.15IRR297 pKa = 11.84AANVLASNRR306 pKa = 11.84ARR308 pKa = 11.84ALRR311 pKa = 11.84RR312 pKa = 11.84EE313 pKa = 4.13IANAGPSTVYY323 pKa = 10.84SKK325 pKa = 9.68MWTKK329 pKa = 10.41VRR331 pKa = 11.84DD332 pKa = 3.98VVWPDD337 pKa = 2.92GALPGFLEE345 pKa = 4.72DD346 pKa = 3.83PGRR349 pKa = 11.84AXGGLSVVHH358 pKa = 6.76GISHH362 pKa = 5.81KK363 pKa = 9.72TGLRR367 pKa = 11.84DD368 pKa = 3.24VRR370 pKa = 11.84ISVDD374 pKa = 2.71RR375 pKa = 11.84HH376 pKa = 4.59ARR378 pKa = 11.84AEE380 pKa = 3.83PRR382 pKa = 11.84PRR384 pKa = 11.84RR385 pKa = 11.84LYY387 pKa = 10.63SIKK390 pKa = 10.22EE391 pKa = 3.66LSANRR396 pKa = 11.84DD397 pKa = 3.42LGVNNADD404 pKa = 3.82INTLEE409 pKa = 4.35CALLEE414 pKa = 3.92RR415 pKa = 11.84MYY417 pKa = 10.3YY418 pKa = 10.36CEE420 pKa = 3.99VGGDD424 pKa = 4.14FVAPPPVDD432 pKa = 3.22PGRR435 pKa = 11.84FTEE438 pKa = 4.24RR439 pKa = 11.84LAGFKK444 pKa = 10.27AALLSNVPEE453 pKa = 4.12PTVMSCEE460 pKa = 4.13EE461 pKa = 4.0VVATYY466 pKa = 10.32SGRR469 pKa = 11.84RR470 pKa = 11.84NTIYY474 pKa = 10.72RR475 pKa = 11.84NALNRR480 pKa = 11.84LTQIGLSRR488 pKa = 11.84SDD490 pKa = 3.05AHH492 pKa = 7.28SIAFVKK498 pKa = 10.19MEE500 pKa = 4.78LVDD503 pKa = 3.39PCKK506 pKa = 10.46APRR509 pKa = 11.84CIQPRR514 pKa = 11.84KK515 pKa = 8.96PVYY518 pKa = 9.73NLCLGRR524 pKa = 11.84FIKK527 pKa = 10.61SVEE530 pKa = 3.47HH531 pKa = 6.99DD532 pKa = 3.3IYY534 pKa = 11.12RR535 pKa = 11.84GIQKK539 pKa = 8.48TFGDD543 pKa = 4.77GPTVMKK549 pKa = 10.2GYY551 pKa = 9.96NVSDD555 pKa = 3.3IGRR558 pKa = 11.84IARR561 pKa = 11.84GKK563 pKa = 8.84WRR565 pKa = 11.84TFKK568 pKa = 11.0KK569 pKa = 10.13PVAIGLDD576 pKa = 3.17AKK578 pKa = 10.7KK579 pKa = 10.7FDD581 pKa = 3.49MHH583 pKa = 7.85VSEE586 pKa = 5.74EE587 pKa = 4.04ALLWEE592 pKa = 4.62HH593 pKa = 6.7GVYY596 pKa = 10.63LDD598 pKa = 5.37LFRR601 pKa = 11.84HH602 pKa = 5.07NPKK605 pKa = 10.33LKK607 pKa = 10.4RR608 pKa = 11.84LLEE611 pKa = 4.03WQVDD615 pKa = 3.57NKK617 pKa = 11.1GAGFCKK623 pKa = 10.25DD624 pKa = 3.02GKK626 pKa = 10.23LKK628 pKa = 10.63YY629 pKa = 9.42RR630 pKa = 11.84VRR632 pKa = 11.84GRR634 pKa = 11.84RR635 pKa = 11.84FSGDD639 pKa = 3.06MNTGLGNCLLMSAMVYY655 pKa = 10.32AYY657 pKa = 10.38AKK659 pKa = 10.55DD660 pKa = 3.29RR661 pKa = 11.84GVGVKK666 pKa = 10.12LLNNGDD672 pKa = 3.98DD673 pKa = 3.9CVVMLEE679 pKa = 4.69SADD682 pKa = 3.65LQQFVTGLDD691 pKa = 3.09EE692 pKa = 4.31WFLEE696 pKa = 3.93MGFRR700 pKa = 11.84MVAEE704 pKa = 4.04EE705 pKa = 4.21PVYY708 pKa = 10.55KK709 pKa = 10.63LHH711 pKa = 7.25QIEE714 pKa = 4.81FCQMHH719 pKa = 7.29PIEE722 pKa = 6.36IGDD725 pKa = 3.79EE726 pKa = 3.92CRR728 pKa = 11.84MVRR731 pKa = 11.84NIPSTLRR738 pKa = 11.84KK739 pKa = 8.88DD740 pKa = 3.58TLTVHH745 pKa = 6.91PLSNQKK751 pKa = 9.46HH752 pKa = 4.82RR753 pKa = 11.84EE754 pKa = 3.94KK755 pKa = 10.18WCTAVGMGGLALTGGVPIMQDD776 pKa = 3.7FYY778 pKa = 11.91QMFQRR783 pKa = 11.84IGCNRR788 pKa = 11.84SSNIANDD795 pKa = 3.58PTFATGLRR803 pKa = 11.84LMSKK807 pKa = 10.31GMAEE811 pKa = 4.89HH812 pKa = 6.39YY813 pKa = 10.05RR814 pKa = 11.84EE815 pKa = 4.37PDD817 pKa = 2.41PWTRR821 pKa = 11.84VQVFEE826 pKa = 4.35AWGITPDD833 pKa = 3.38EE834 pKa = 4.08QKK836 pKa = 10.93AIEE839 pKa = 4.31RR840 pKa = 11.84YY841 pKa = 9.55YY842 pKa = 11.15RR843 pKa = 11.84DD844 pKa = 3.76FSLSDD849 pKa = 3.3QPVSGEE855 pKa = 3.87QANVRR860 pKa = 11.84CHH862 pKa = 7.38LLLQLL867 pKa = 4.53
MM1 pKa = 7.64SFFEE5 pKa = 4.45LLRR8 pKa = 11.84AFLCCATADD17 pKa = 3.75SGEE20 pKa = 4.08VKK22 pKa = 10.0MDD24 pKa = 3.33VAKK27 pKa = 9.89TLLRR31 pKa = 11.84EE32 pKa = 4.33DD33 pKa = 4.77CLATDD38 pKa = 4.49NIEE41 pKa = 4.1FTHH44 pKa = 6.87DD45 pKa = 3.63PKK47 pKa = 10.59EE48 pKa = 3.95HH49 pKa = 6.21HH50 pKa = 5.52VTRR53 pKa = 11.84RR54 pKa = 11.84PAGVRR59 pKa = 11.84VTDD62 pKa = 3.49NRR64 pKa = 11.84EE65 pKa = 3.61ISPLEE70 pKa = 3.88PRR72 pKa = 11.84AGLPVKK78 pKa = 10.18KK79 pKa = 9.8CAYY82 pKa = 8.25RR83 pKa = 11.84AEE85 pKa = 4.1MDD87 pKa = 3.57VLRR90 pKa = 11.84KK91 pKa = 9.75LRR93 pKa = 11.84LAAEE97 pKa = 4.37DD98 pKa = 3.44AFGRR102 pKa = 11.84QQEE105 pKa = 4.41KK106 pKa = 10.24KK107 pKa = 10.17QPDD110 pKa = 4.03PVTQHH115 pKa = 7.02RR116 pKa = 11.84LEE118 pKa = 4.42CVLNKK123 pKa = 9.46ATLAAFKK130 pKa = 10.92VVGATHH136 pKa = 5.86QCEE139 pKa = 4.32SWSLLVAIGEE149 pKa = 4.31LQLDD153 pKa = 3.87MTHH156 pKa = 6.65TRR158 pKa = 11.84SEE160 pKa = 3.85WKK162 pKa = 10.05RR163 pKa = 11.84RR164 pKa = 11.84LLRR167 pKa = 11.84LGEE170 pKa = 4.16LCSIEE175 pKa = 5.15SGCEE179 pKa = 3.59WEE181 pKa = 5.15VEE183 pKa = 4.3GNPPGLASEE192 pKa = 4.13PHH194 pKa = 6.37PPSGEE199 pKa = 4.06VPVPTDD205 pKa = 2.99QPTGVHH211 pKa = 6.68RR212 pKa = 11.84LCHH215 pKa = 6.11DD216 pKa = 3.61VVEE219 pKa = 5.18VKK221 pKa = 10.19AHH223 pKa = 4.96RR224 pKa = 11.84RR225 pKa = 11.84IRR227 pKa = 11.84KK228 pKa = 9.11DD229 pKa = 3.03RR230 pKa = 11.84EE231 pKa = 3.85GQYY234 pKa = 10.09VGTVVRR240 pKa = 11.84EE241 pKa = 3.8LKK243 pKa = 10.81NRR245 pKa = 11.84FGCPKK250 pKa = 8.82PTEE253 pKa = 4.56DD254 pKa = 3.55NLLAVRR260 pKa = 11.84RR261 pKa = 11.84SAINIMEE268 pKa = 4.16KK269 pKa = 10.22HH270 pKa = 5.65GVRR273 pKa = 11.84PSHH276 pKa = 6.77VGEE279 pKa = 4.2VVDD282 pKa = 4.98LVVAGVFVPDD292 pKa = 3.53RR293 pKa = 11.84NEE295 pKa = 3.15IRR297 pKa = 11.84AANVLASNRR306 pKa = 11.84ARR308 pKa = 11.84ALRR311 pKa = 11.84RR312 pKa = 11.84EE313 pKa = 4.13IANAGPSTVYY323 pKa = 10.84SKK325 pKa = 9.68MWTKK329 pKa = 10.41VRR331 pKa = 11.84DD332 pKa = 3.98VVWPDD337 pKa = 2.92GALPGFLEE345 pKa = 4.72DD346 pKa = 3.83PGRR349 pKa = 11.84AXGGLSVVHH358 pKa = 6.76GISHH362 pKa = 5.81KK363 pKa = 9.72TGLRR367 pKa = 11.84DD368 pKa = 3.24VRR370 pKa = 11.84ISVDD374 pKa = 2.71RR375 pKa = 11.84HH376 pKa = 4.59ARR378 pKa = 11.84AEE380 pKa = 3.83PRR382 pKa = 11.84PRR384 pKa = 11.84RR385 pKa = 11.84LYY387 pKa = 10.63SIKK390 pKa = 10.22EE391 pKa = 3.66LSANRR396 pKa = 11.84DD397 pKa = 3.42LGVNNADD404 pKa = 3.82INTLEE409 pKa = 4.35CALLEE414 pKa = 3.92RR415 pKa = 11.84MYY417 pKa = 10.3YY418 pKa = 10.36CEE420 pKa = 3.99VGGDD424 pKa = 4.14FVAPPPVDD432 pKa = 3.22PGRR435 pKa = 11.84FTEE438 pKa = 4.24RR439 pKa = 11.84LAGFKK444 pKa = 10.27AALLSNVPEE453 pKa = 4.12PTVMSCEE460 pKa = 4.13EE461 pKa = 4.0VVATYY466 pKa = 10.32SGRR469 pKa = 11.84RR470 pKa = 11.84NTIYY474 pKa = 10.72RR475 pKa = 11.84NALNRR480 pKa = 11.84LTQIGLSRR488 pKa = 11.84SDD490 pKa = 3.05AHH492 pKa = 7.28SIAFVKK498 pKa = 10.19MEE500 pKa = 4.78LVDD503 pKa = 3.39PCKK506 pKa = 10.46APRR509 pKa = 11.84CIQPRR514 pKa = 11.84KK515 pKa = 8.96PVYY518 pKa = 9.73NLCLGRR524 pKa = 11.84FIKK527 pKa = 10.61SVEE530 pKa = 3.47HH531 pKa = 6.99DD532 pKa = 3.3IYY534 pKa = 11.12RR535 pKa = 11.84GIQKK539 pKa = 8.48TFGDD543 pKa = 4.77GPTVMKK549 pKa = 10.2GYY551 pKa = 9.96NVSDD555 pKa = 3.3IGRR558 pKa = 11.84IARR561 pKa = 11.84GKK563 pKa = 8.84WRR565 pKa = 11.84TFKK568 pKa = 11.0KK569 pKa = 10.13PVAIGLDD576 pKa = 3.17AKK578 pKa = 10.7KK579 pKa = 10.7FDD581 pKa = 3.49MHH583 pKa = 7.85VSEE586 pKa = 5.74EE587 pKa = 4.04ALLWEE592 pKa = 4.62HH593 pKa = 6.7GVYY596 pKa = 10.63LDD598 pKa = 5.37LFRR601 pKa = 11.84HH602 pKa = 5.07NPKK605 pKa = 10.33LKK607 pKa = 10.4RR608 pKa = 11.84LLEE611 pKa = 4.03WQVDD615 pKa = 3.57NKK617 pKa = 11.1GAGFCKK623 pKa = 10.25DD624 pKa = 3.02GKK626 pKa = 10.23LKK628 pKa = 10.63YY629 pKa = 9.42RR630 pKa = 11.84VRR632 pKa = 11.84GRR634 pKa = 11.84RR635 pKa = 11.84FSGDD639 pKa = 3.06MNTGLGNCLLMSAMVYY655 pKa = 10.32AYY657 pKa = 10.38AKK659 pKa = 10.55DD660 pKa = 3.29RR661 pKa = 11.84GVGVKK666 pKa = 10.12LLNNGDD672 pKa = 3.98DD673 pKa = 3.9CVVMLEE679 pKa = 4.69SADD682 pKa = 3.65LQQFVTGLDD691 pKa = 3.09EE692 pKa = 4.31WFLEE696 pKa = 3.93MGFRR700 pKa = 11.84MVAEE704 pKa = 4.04EE705 pKa = 4.21PVYY708 pKa = 10.55KK709 pKa = 10.63LHH711 pKa = 7.25QIEE714 pKa = 4.81FCQMHH719 pKa = 7.29PIEE722 pKa = 6.36IGDD725 pKa = 3.79EE726 pKa = 3.92CRR728 pKa = 11.84MVRR731 pKa = 11.84NIPSTLRR738 pKa = 11.84KK739 pKa = 8.88DD740 pKa = 3.58TLTVHH745 pKa = 6.91PLSNQKK751 pKa = 9.46HH752 pKa = 4.82RR753 pKa = 11.84EE754 pKa = 3.94KK755 pKa = 10.18WCTAVGMGGLALTGGVPIMQDD776 pKa = 3.7FYY778 pKa = 11.91QMFQRR783 pKa = 11.84IGCNRR788 pKa = 11.84SSNIANDD795 pKa = 3.58PTFATGLRR803 pKa = 11.84LMSKK807 pKa = 10.31GMAEE811 pKa = 4.89HH812 pKa = 6.39YY813 pKa = 10.05RR814 pKa = 11.84EE815 pKa = 4.37PDD817 pKa = 2.41PWTRR821 pKa = 11.84VQVFEE826 pKa = 4.35AWGITPDD833 pKa = 3.38EE834 pKa = 4.08QKK836 pKa = 10.93AIEE839 pKa = 4.31RR840 pKa = 11.84YY841 pKa = 9.55YY842 pKa = 11.15RR843 pKa = 11.84DD844 pKa = 3.76FSLSDD849 pKa = 3.3QPVSGEE855 pKa = 3.87QANVRR860 pKa = 11.84CHH862 pKa = 7.38LLLQLL867 pKa = 4.53
Molecular weight: 97.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFV5|A0A1L3KFV5_9VIRU Capsid protein OS=Changjiang tombus-like virus 9 OX=1922823 PE=3 SV=1
MM1 pKa = 7.44PPVVAAVKK9 pKa = 10.39AGEE12 pKa = 4.31LRR14 pKa = 11.84QQKK17 pKa = 10.74YY18 pKa = 10.88NMVKK22 pKa = 10.35KK23 pKa = 10.56ANKK26 pKa = 9.43KK27 pKa = 7.4MKK29 pKa = 9.21KK30 pKa = 7.63TIKK33 pKa = 9.28VAKK36 pKa = 8.93KK37 pKa = 9.46KK38 pKa = 11.05SEE40 pKa = 3.65ITEE43 pKa = 4.06LGRR46 pKa = 11.84VLRR49 pKa = 11.84TLGGIGGGFAGGMIGQGAAGSAVGTGLGAAVSRR82 pKa = 11.84WLGQGDD88 pKa = 4.02YY89 pKa = 10.89SVTSNSVLRR98 pKa = 11.84AATTVPSMHH107 pKa = 7.02KK108 pKa = 10.03NDD110 pKa = 3.34QTVVVRR116 pKa = 11.84HH117 pKa = 5.82KK118 pKa = 10.87EE119 pKa = 3.87FVGEE123 pKa = 4.23LRR125 pKa = 11.84GNQNFTVAYY134 pKa = 9.41RR135 pKa = 11.84YY136 pKa = 10.34SLNPGIALTFPWLHH150 pKa = 6.71SIAAQYY156 pKa = 10.38SEE158 pKa = 3.69YY159 pKa = 10.21RR160 pKa = 11.84IRR162 pKa = 11.84GMVFHH167 pKa = 6.44YY168 pKa = 10.68VPTSGASVASSNTALGSVMFQTSYY192 pKa = 10.67RR193 pKa = 11.84ASEE196 pKa = 4.15NPPGSKK202 pKa = 8.86IEE204 pKa = 3.93MMNEE208 pKa = 3.32YY209 pKa = 8.79WASEE213 pKa = 3.76GRR215 pKa = 11.84PCDD218 pKa = 3.97EE219 pKa = 4.11FCHH222 pKa = 6.84PIEE225 pKa = 5.36CDD227 pKa = 3.21PKK229 pKa = 9.53EE230 pKa = 4.09NPFNVQYY237 pKa = 11.03VRR239 pKa = 11.84TGGLPAGEE247 pKa = 4.2NLLMYY252 pKa = 10.55DD253 pKa = 4.45LGTTTVAVTGMQTTGTVLGDD273 pKa = 3.17IWCTYY278 pKa = 7.91EE279 pKa = 5.08VEE281 pKa = 4.24LKK283 pKa = 10.57KK284 pKa = 10.68PKK286 pKa = 9.33LTGLNTEE293 pKa = 4.45ATRR296 pKa = 11.84SLYY299 pKa = 11.05SNSSTGITNANPLGSGPVLSTIDD322 pKa = 3.4GVSAVGSQITFPGNLDD338 pKa = 3.19GKK340 pKa = 9.27YY341 pKa = 10.07VVVLTISGFTAAGGVTVMTSGPATLTSLAQAQGSASLSYY380 pKa = 10.7VAILEE385 pKa = 4.31FTGSVNTTLLTISLTTLTGATLSTLRR411 pKa = 11.84ITEE414 pKa = 4.06WNADD418 pKa = 3.66FAA420 pKa = 6.14
MM1 pKa = 7.44PPVVAAVKK9 pKa = 10.39AGEE12 pKa = 4.31LRR14 pKa = 11.84QQKK17 pKa = 10.74YY18 pKa = 10.88NMVKK22 pKa = 10.35KK23 pKa = 10.56ANKK26 pKa = 9.43KK27 pKa = 7.4MKK29 pKa = 9.21KK30 pKa = 7.63TIKK33 pKa = 9.28VAKK36 pKa = 8.93KK37 pKa = 9.46KK38 pKa = 11.05SEE40 pKa = 3.65ITEE43 pKa = 4.06LGRR46 pKa = 11.84VLRR49 pKa = 11.84TLGGIGGGFAGGMIGQGAAGSAVGTGLGAAVSRR82 pKa = 11.84WLGQGDD88 pKa = 4.02YY89 pKa = 10.89SVTSNSVLRR98 pKa = 11.84AATTVPSMHH107 pKa = 7.02KK108 pKa = 10.03NDD110 pKa = 3.34QTVVVRR116 pKa = 11.84HH117 pKa = 5.82KK118 pKa = 10.87EE119 pKa = 3.87FVGEE123 pKa = 4.23LRR125 pKa = 11.84GNQNFTVAYY134 pKa = 9.41RR135 pKa = 11.84YY136 pKa = 10.34SLNPGIALTFPWLHH150 pKa = 6.71SIAAQYY156 pKa = 10.38SEE158 pKa = 3.69YY159 pKa = 10.21RR160 pKa = 11.84IRR162 pKa = 11.84GMVFHH167 pKa = 6.44YY168 pKa = 10.68VPTSGASVASSNTALGSVMFQTSYY192 pKa = 10.67RR193 pKa = 11.84ASEE196 pKa = 4.15NPPGSKK202 pKa = 8.86IEE204 pKa = 3.93MMNEE208 pKa = 3.32YY209 pKa = 8.79WASEE213 pKa = 3.76GRR215 pKa = 11.84PCDD218 pKa = 3.97EE219 pKa = 4.11FCHH222 pKa = 6.84PIEE225 pKa = 5.36CDD227 pKa = 3.21PKK229 pKa = 9.53EE230 pKa = 4.09NPFNVQYY237 pKa = 11.03VRR239 pKa = 11.84TGGLPAGEE247 pKa = 4.2NLLMYY252 pKa = 10.55DD253 pKa = 4.45LGTTTVAVTGMQTTGTVLGDD273 pKa = 3.17IWCTYY278 pKa = 7.91EE279 pKa = 5.08VEE281 pKa = 4.24LKK283 pKa = 10.57KK284 pKa = 10.68PKK286 pKa = 9.33LTGLNTEE293 pKa = 4.45ATRR296 pKa = 11.84SLYY299 pKa = 11.05SNSSTGITNANPLGSGPVLSTIDD322 pKa = 3.4GVSAVGSQITFPGNLDD338 pKa = 3.19GKK340 pKa = 9.27YY341 pKa = 10.07VVVLTISGFTAAGGVTVMTSGPATLTSLAQAQGSASLSYY380 pKa = 10.7VAILEE385 pKa = 4.31FTGSVNTTLLTISLTTLTGATLSTLRR411 pKa = 11.84ITEE414 pKa = 4.06WNADD418 pKa = 3.66FAA420 pKa = 6.14
Molecular weight: 44.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1287 |
420 |
867 |
643.5 |
70.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.77 ± 0.554 | 2.176 ± 0.652 |
4.507 ± 1.261 | 6.138 ± 0.861 |
3.186 ± 0.175 | 8.702 ± 1.327 |
2.409 ± 0.65 | 3.807 ± 0.128 |
5.128 ± 0.195 | 8.936 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.797 ± 0.032 | 4.118 ± 0.343 |
5.128 ± 0.449 | 2.953 ± 0.076 |
7.148 ± 1.908 | 5.905 ± 1.422 |
6.449 ± 2.275 | 8.625 ± 0.028 |
1.321 ± 0.07 | 2.72 ± 0.454 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
