
Arumowot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Arumowot phlebovirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
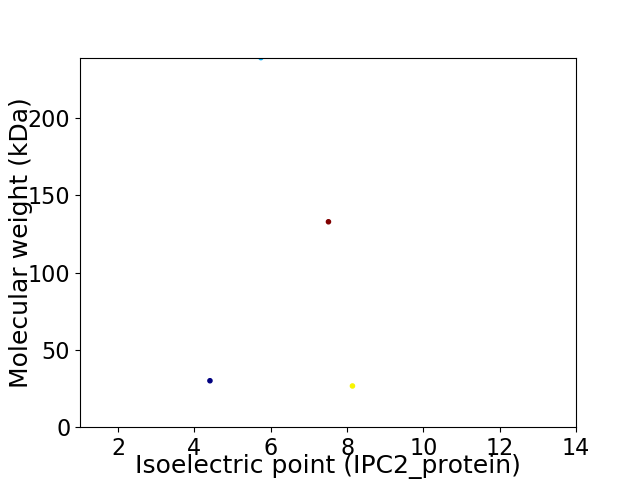
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1SV58|I1SV58_9VIRU Nucleoprotein OS=Arumowot virus OX=904698 PE=3 SV=1
MM1 pKa = 7.83ASPYY5 pKa = 10.96NYY7 pKa = 9.59IHH9 pKa = 7.72DD10 pKa = 4.53LVTPNYY16 pKa = 9.42NGPIPGEE23 pKa = 4.13VVVSFEE29 pKa = 4.28PVNSYY34 pKa = 10.71VNNPVCVYY42 pKa = 10.49EE43 pKa = 4.43DD44 pKa = 4.07LEE46 pKa = 4.44FEE48 pKa = 4.05ISGMGLCTMTSEE60 pKa = 4.54SLYY63 pKa = 11.24DD64 pKa = 3.93FLDD67 pKa = 3.88SGLMPWRR74 pKa = 11.84WGKK77 pKa = 11.12GMFEE81 pKa = 4.23SRR83 pKa = 11.84LTAMEE88 pKa = 4.36TPLFDD93 pKa = 5.17SFLKK97 pKa = 9.52QIPMIPTTQLFSVGRR112 pKa = 11.84DD113 pKa = 3.51PLRR116 pKa = 11.84AALSWPTGRR125 pKa = 11.84CSLRR129 pKa = 11.84FFRR132 pKa = 11.84QQWGSDD138 pKa = 2.87AMSYY142 pKa = 10.26QSNRR146 pKa = 11.84KK147 pKa = 8.78LRR149 pKa = 11.84ARR151 pKa = 11.84LLFQSTQSSDD161 pKa = 3.41LEE163 pKa = 4.23TAIITLAKK171 pKa = 9.83DD172 pKa = 3.11IRR174 pKa = 11.84GMSSILDD181 pKa = 3.65FPLNLIPGFDD191 pKa = 3.84LLKK194 pKa = 10.56CVSILQFIRR203 pKa = 11.84ALRR206 pKa = 11.84SYY208 pKa = 10.72PEE210 pKa = 4.12TEE212 pKa = 5.04AEE214 pKa = 4.07RR215 pKa = 11.84QSNGLRR221 pKa = 11.84DD222 pKa = 4.02DD223 pKa = 4.74LDD225 pKa = 3.72IFLGSARR232 pKa = 11.84EE233 pKa = 3.91ALSRR237 pKa = 11.84DD238 pKa = 3.71FPALIPAITPEE249 pKa = 4.39SDD251 pKa = 3.27DD252 pKa = 4.24SLWDD256 pKa = 3.63SGSSCEE262 pKa = 5.43DD263 pKa = 3.49DD264 pKa = 3.73VDD266 pKa = 3.79VV267 pKa = 5.14
MM1 pKa = 7.83ASPYY5 pKa = 10.96NYY7 pKa = 9.59IHH9 pKa = 7.72DD10 pKa = 4.53LVTPNYY16 pKa = 9.42NGPIPGEE23 pKa = 4.13VVVSFEE29 pKa = 4.28PVNSYY34 pKa = 10.71VNNPVCVYY42 pKa = 10.49EE43 pKa = 4.43DD44 pKa = 4.07LEE46 pKa = 4.44FEE48 pKa = 4.05ISGMGLCTMTSEE60 pKa = 4.54SLYY63 pKa = 11.24DD64 pKa = 3.93FLDD67 pKa = 3.88SGLMPWRR74 pKa = 11.84WGKK77 pKa = 11.12GMFEE81 pKa = 4.23SRR83 pKa = 11.84LTAMEE88 pKa = 4.36TPLFDD93 pKa = 5.17SFLKK97 pKa = 9.52QIPMIPTTQLFSVGRR112 pKa = 11.84DD113 pKa = 3.51PLRR116 pKa = 11.84AALSWPTGRR125 pKa = 11.84CSLRR129 pKa = 11.84FFRR132 pKa = 11.84QQWGSDD138 pKa = 2.87AMSYY142 pKa = 10.26QSNRR146 pKa = 11.84KK147 pKa = 8.78LRR149 pKa = 11.84ARR151 pKa = 11.84LLFQSTQSSDD161 pKa = 3.41LEE163 pKa = 4.23TAIITLAKK171 pKa = 9.83DD172 pKa = 3.11IRR174 pKa = 11.84GMSSILDD181 pKa = 3.65FPLNLIPGFDD191 pKa = 3.84LLKK194 pKa = 10.56CVSILQFIRR203 pKa = 11.84ALRR206 pKa = 11.84SYY208 pKa = 10.72PEE210 pKa = 4.12TEE212 pKa = 5.04AEE214 pKa = 4.07RR215 pKa = 11.84QSNGLRR221 pKa = 11.84DD222 pKa = 4.02DD223 pKa = 4.74LDD225 pKa = 3.72IFLGSARR232 pKa = 11.84EE233 pKa = 3.91ALSRR237 pKa = 11.84DD238 pKa = 3.71FPALIPAITPEE249 pKa = 4.39SDD251 pKa = 3.27DD252 pKa = 4.24SLWDD256 pKa = 3.63SGSSCEE262 pKa = 5.43DD263 pKa = 3.49DD264 pKa = 3.73VDD266 pKa = 3.79VV267 pKa = 5.14
Molecular weight: 30.05 kDa
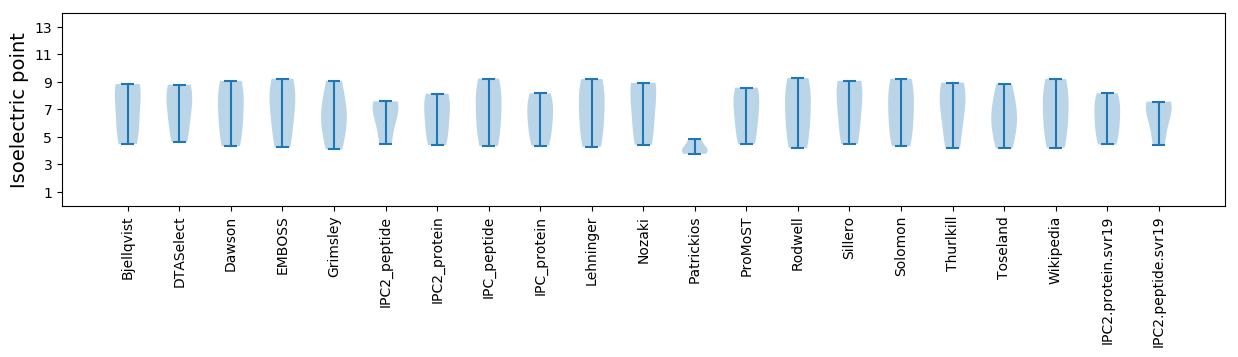
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1SV58|I1SV58_9VIRU Nucleoprotein OS=Arumowot virus OX=904698 PE=3 SV=1
MM1 pKa = 8.0DD2 pKa = 5.09NYY4 pKa = 11.43AKK6 pKa = 10.33IAIDD10 pKa = 3.69FAGDD14 pKa = 3.87AIDD17 pKa = 3.96ASEE20 pKa = 4.94LIALAEE26 pKa = 4.01EE27 pKa = 4.93FSFNGFDD34 pKa = 3.8PNAIMKK40 pKa = 9.1LIIEE44 pKa = 4.37RR45 pKa = 11.84GGNNWKK51 pKa = 10.14EE52 pKa = 4.11DD53 pKa = 3.77AKK55 pKa = 10.67MMIVIGITRR64 pKa = 11.84GNKK67 pKa = 7.77IKK69 pKa = 11.12KK70 pKa = 9.03MMDD73 pKa = 2.75RR74 pKa = 11.84MGEE77 pKa = 3.91EE78 pKa = 4.5GKK80 pKa = 8.68TKK82 pKa = 10.67LKK84 pKa = 10.9YY85 pKa = 10.61LVDD88 pKa = 3.65KK89 pKa = 11.09YY90 pKa = 10.9KK91 pKa = 10.93LKK93 pKa = 10.34EE94 pKa = 4.0ASPGARR100 pKa = 11.84DD101 pKa = 3.5LTLSRR106 pKa = 11.84VAAVLAPWTVQALRR120 pKa = 11.84VLEE123 pKa = 4.26NKK125 pKa = 10.51LPVTGATMDD134 pKa = 4.16SLSHH138 pKa = 6.41NYY140 pKa = 8.57PRR142 pKa = 11.84ALMHH146 pKa = 6.33SAAGGILDD154 pKa = 4.78PGLGDD159 pKa = 3.54ACFGALLQAHH169 pKa = 6.41SLYY172 pKa = 10.85LDD174 pKa = 3.43TFSRR178 pKa = 11.84LINPPLRR185 pKa = 11.84SKK187 pKa = 10.8SKK189 pKa = 10.89QEE191 pKa = 3.78VAQSFSSALQAAYY204 pKa = 10.36SSNFFSPAQKK214 pKa = 10.31RR215 pKa = 11.84QILQTMGLIDD225 pKa = 4.78MNLQPTKK232 pKa = 10.8AVLDD236 pKa = 3.58AAGIYY241 pKa = 10.95NSMM244 pKa = 4.28
MM1 pKa = 8.0DD2 pKa = 5.09NYY4 pKa = 11.43AKK6 pKa = 10.33IAIDD10 pKa = 3.69FAGDD14 pKa = 3.87AIDD17 pKa = 3.96ASEE20 pKa = 4.94LIALAEE26 pKa = 4.01EE27 pKa = 4.93FSFNGFDD34 pKa = 3.8PNAIMKK40 pKa = 9.1LIIEE44 pKa = 4.37RR45 pKa = 11.84GGNNWKK51 pKa = 10.14EE52 pKa = 4.11DD53 pKa = 3.77AKK55 pKa = 10.67MMIVIGITRR64 pKa = 11.84GNKK67 pKa = 7.77IKK69 pKa = 11.12KK70 pKa = 9.03MMDD73 pKa = 2.75RR74 pKa = 11.84MGEE77 pKa = 3.91EE78 pKa = 4.5GKK80 pKa = 8.68TKK82 pKa = 10.67LKK84 pKa = 10.9YY85 pKa = 10.61LVDD88 pKa = 3.65KK89 pKa = 11.09YY90 pKa = 10.9KK91 pKa = 10.93LKK93 pKa = 10.34EE94 pKa = 4.0ASPGARR100 pKa = 11.84DD101 pKa = 3.5LTLSRR106 pKa = 11.84VAAVLAPWTVQALRR120 pKa = 11.84VLEE123 pKa = 4.26NKK125 pKa = 10.51LPVTGATMDD134 pKa = 4.16SLSHH138 pKa = 6.41NYY140 pKa = 8.57PRR142 pKa = 11.84ALMHH146 pKa = 6.33SAAGGILDD154 pKa = 4.78PGLGDD159 pKa = 3.54ACFGALLQAHH169 pKa = 6.41SLYY172 pKa = 10.85LDD174 pKa = 3.43TFSRR178 pKa = 11.84LINPPLRR185 pKa = 11.84SKK187 pKa = 10.8SKK189 pKa = 10.89QEE191 pKa = 3.78VAQSFSSALQAAYY204 pKa = 10.36SSNFFSPAQKK214 pKa = 10.31RR215 pKa = 11.84QILQTMGLIDD225 pKa = 4.78MNLQPTKK232 pKa = 10.8AVLDD236 pKa = 3.58AAGIYY241 pKa = 10.95NSMM244 pKa = 4.28
Molecular weight: 26.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3814 |
244 |
2093 |
953.5 |
107.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.929 ± 1.267 | 2.727 ± 0.878 |
5.716 ± 0.805 | 6.266 ± 0.644 |
4.274 ± 0.413 | 6.162 ± 0.498 |
1.966 ± 0.314 | 6.66 ± 0.224 |
6.607 ± 0.713 | 9.439 ± 0.666 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.543 ± 0.609 | 4.143 ± 0.169 |
4.064 ± 0.401 | 3.54 ± 0.234 |
4.929 ± 0.743 | 9.622 ± 0.516 |
5.821 ± 0.564 | 6.135 ± 0.624 |
1.337 ± 0.119 | 3.12 ± 0.098 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
