
Blautia sp. OF03-15BH
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Blautia; unclassified Blautia
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
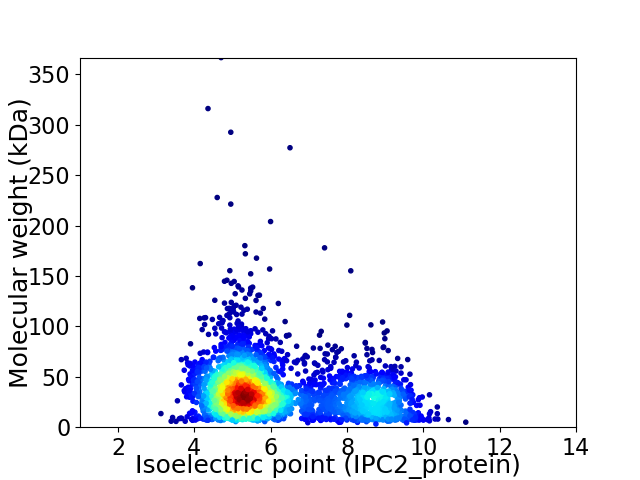
Virtual 2D-PAGE plot for 3234 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A413I0U7|A0A413I0U7_9FIRM Phosphotyrosine protein phosphatase OS=Blautia sp. OF03-15BH OX=2292287 GN=DXA57_10265 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.43KK3 pKa = 8.69FTKK6 pKa = 10.03FVSAVAAVSLVAAPMAVSADD26 pKa = 3.31EE27 pKa = 4.23KK28 pKa = 10.53TFLLGGSGPLTGAAASYY45 pKa = 7.46GTSVKK50 pKa = 10.29QGAEE54 pKa = 3.58IAIQEE59 pKa = 4.15INDD62 pKa = 3.61AGGAQVGEE70 pKa = 4.12DD71 pKa = 3.66TYY73 pKa = 11.79KK74 pKa = 11.14LEE76 pKa = 5.14LAFEE80 pKa = 4.85DD81 pKa = 5.72DD82 pKa = 4.23EE83 pKa = 4.4ATEE86 pKa = 4.41DD87 pKa = 4.67KK88 pKa = 10.95AVQAYY93 pKa = 6.98NTLMDD98 pKa = 3.7KK99 pKa = 10.97GINGFLGCTTSGSCIAVAQLSQQDD123 pKa = 4.63GILQITPSGSAEE135 pKa = 4.02GCIEE139 pKa = 4.19PDD141 pKa = 2.93NAFRR145 pKa = 11.84ICFSDD150 pKa = 3.83PEE152 pKa = 4.27QGVAMADD159 pKa = 3.67YY160 pKa = 11.1AVDD163 pKa = 3.26TLGYY167 pKa = 10.2KK168 pKa = 10.23KK169 pKa = 10.2IAVLYY174 pKa = 9.8NVADD178 pKa = 4.43EE179 pKa = 4.49YY180 pKa = 11.33STGIYY185 pKa = 10.33DD186 pKa = 4.18AFTAEE191 pKa = 4.23IEE193 pKa = 4.33AKK195 pKa = 10.21GAEE198 pKa = 4.18IVDD201 pKa = 3.44AEE203 pKa = 4.44SFNTGDD209 pKa = 3.55VDD211 pKa = 4.84FNSQLTSIKK220 pKa = 9.79GTDD223 pKa = 3.1AEE225 pKa = 4.72AIYY228 pKa = 10.81VPAYY232 pKa = 9.41YY233 pKa = 10.24QDD235 pKa = 2.66ITYY238 pKa = 8.55ITKK241 pKa = 9.85QASEE245 pKa = 4.79LGMEE249 pKa = 4.45LPFLGSDD256 pKa = 2.99GWDD259 pKa = 3.16GVLKK263 pKa = 9.96TVTDD267 pKa = 3.84PATVEE272 pKa = 4.14GCIFTSPFCASVDD285 pKa = 3.64DD286 pKa = 4.36EE287 pKa = 4.5KK288 pKa = 11.67VVNFVKK294 pKa = 10.49AYY296 pKa = 8.56QDD298 pKa = 3.58AYY300 pKa = 10.98GEE302 pKa = 4.48TPDD305 pKa = 4.65QFAADD310 pKa = 4.33AYY312 pKa = 10.64DD313 pKa = 3.29GVYY316 pKa = 10.64AFKK319 pKa = 10.84AAMEE323 pKa = 4.14EE324 pKa = 4.04AGSIEE329 pKa = 4.26SADD332 pKa = 4.38MIDD335 pKa = 4.19AMTKK339 pKa = 9.75ISVSGLTGDD348 pKa = 5.53PITFDD353 pKa = 3.24ASGAAVKK360 pKa = 10.06DD361 pKa = 3.87VKK363 pKa = 10.73FVTVKK368 pKa = 10.53DD369 pKa = 3.69GQYY372 pKa = 10.31TYY374 pKa = 11.59VEE376 pKa = 4.21
MM1 pKa = 7.55KK2 pKa = 10.43KK3 pKa = 8.69FTKK6 pKa = 10.03FVSAVAAVSLVAAPMAVSADD26 pKa = 3.31EE27 pKa = 4.23KK28 pKa = 10.53TFLLGGSGPLTGAAASYY45 pKa = 7.46GTSVKK50 pKa = 10.29QGAEE54 pKa = 3.58IAIQEE59 pKa = 4.15INDD62 pKa = 3.61AGGAQVGEE70 pKa = 4.12DD71 pKa = 3.66TYY73 pKa = 11.79KK74 pKa = 11.14LEE76 pKa = 5.14LAFEE80 pKa = 4.85DD81 pKa = 5.72DD82 pKa = 4.23EE83 pKa = 4.4ATEE86 pKa = 4.41DD87 pKa = 4.67KK88 pKa = 10.95AVQAYY93 pKa = 6.98NTLMDD98 pKa = 3.7KK99 pKa = 10.97GINGFLGCTTSGSCIAVAQLSQQDD123 pKa = 4.63GILQITPSGSAEE135 pKa = 4.02GCIEE139 pKa = 4.19PDD141 pKa = 2.93NAFRR145 pKa = 11.84ICFSDD150 pKa = 3.83PEE152 pKa = 4.27QGVAMADD159 pKa = 3.67YY160 pKa = 11.1AVDD163 pKa = 3.26TLGYY167 pKa = 10.2KK168 pKa = 10.23KK169 pKa = 10.2IAVLYY174 pKa = 9.8NVADD178 pKa = 4.43EE179 pKa = 4.49YY180 pKa = 11.33STGIYY185 pKa = 10.33DD186 pKa = 4.18AFTAEE191 pKa = 4.23IEE193 pKa = 4.33AKK195 pKa = 10.21GAEE198 pKa = 4.18IVDD201 pKa = 3.44AEE203 pKa = 4.44SFNTGDD209 pKa = 3.55VDD211 pKa = 4.84FNSQLTSIKK220 pKa = 9.79GTDD223 pKa = 3.1AEE225 pKa = 4.72AIYY228 pKa = 10.81VPAYY232 pKa = 9.41YY233 pKa = 10.24QDD235 pKa = 2.66ITYY238 pKa = 8.55ITKK241 pKa = 9.85QASEE245 pKa = 4.79LGMEE249 pKa = 4.45LPFLGSDD256 pKa = 2.99GWDD259 pKa = 3.16GVLKK263 pKa = 9.96TVTDD267 pKa = 3.84PATVEE272 pKa = 4.14GCIFTSPFCASVDD285 pKa = 3.64DD286 pKa = 4.36EE287 pKa = 4.5KK288 pKa = 11.67VVNFVKK294 pKa = 10.49AYY296 pKa = 8.56QDD298 pKa = 3.58AYY300 pKa = 10.98GEE302 pKa = 4.48TPDD305 pKa = 4.65QFAADD310 pKa = 4.33AYY312 pKa = 10.64DD313 pKa = 3.29GVYY316 pKa = 10.64AFKK319 pKa = 10.84AAMEE323 pKa = 4.14EE324 pKa = 4.04AGSIEE329 pKa = 4.26SADD332 pKa = 4.38MIDD335 pKa = 4.19AMTKK339 pKa = 9.75ISVSGLTGDD348 pKa = 5.53PITFDD353 pKa = 3.24ASGAAVKK360 pKa = 10.06DD361 pKa = 3.87VKK363 pKa = 10.73FVTVKK368 pKa = 10.53DD369 pKa = 3.69GQYY372 pKa = 10.31TYY374 pKa = 11.59VEE376 pKa = 4.21
Molecular weight: 39.62 kDa
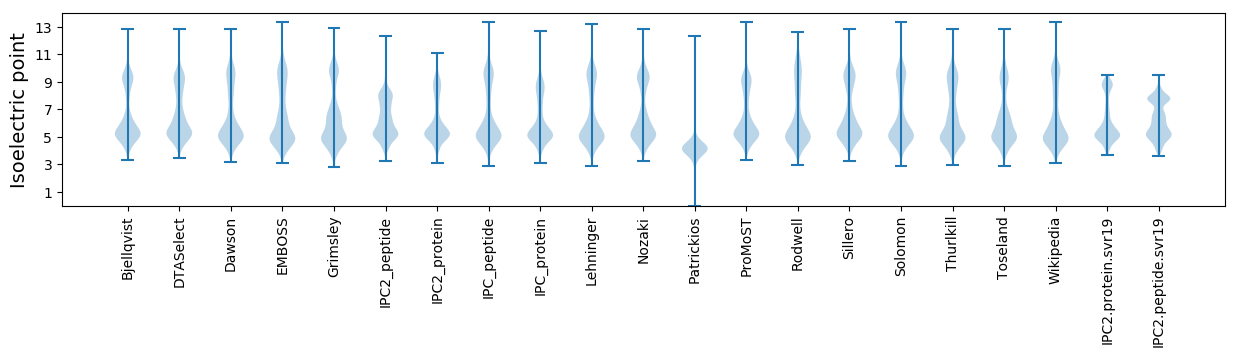
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A413HYF6|A0A413HYF6_9FIRM Rhodanese-like domain-containing protein OS=Blautia sp. OF03-15BH OX=2292287 GN=DXA57_14260 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1085421 |
27 |
3430 |
335.6 |
37.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.442 ± 0.043 | 1.472 ± 0.018 |
5.449 ± 0.034 | 8.076 ± 0.066 |
4.222 ± 0.035 | 7.152 ± 0.039 |
1.772 ± 0.017 | 6.683 ± 0.048 |
6.827 ± 0.044 | 9.4 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.962 ± 0.023 | 4.074 ± 0.029 |
3.465 ± 0.029 | 3.186 ± 0.024 |
4.657 ± 0.037 | 5.801 ± 0.036 |
5.472 ± 0.044 | 6.727 ± 0.034 |
1.021 ± 0.014 | 4.141 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
