
Paraburkholderia caballeronis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
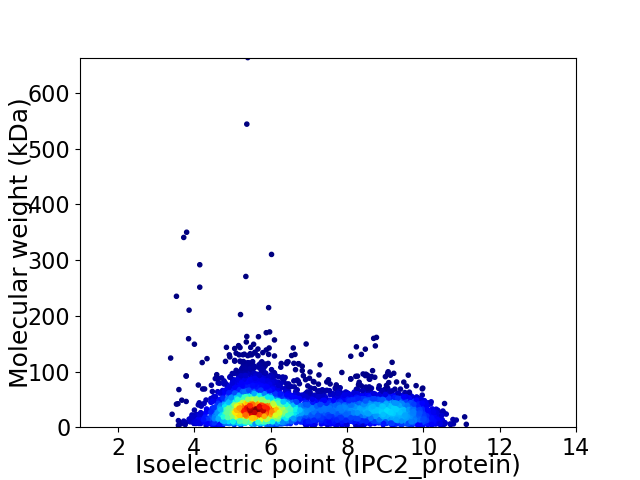
Virtual 2D-PAGE plot for 6189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7UIR0|A0A1H7UIR0_9BURK Probable 2-(5''-triphosphoribosyl)-3'-dephosphocoenzyme-A synthase OS=Paraburkholderia caballeronis OX=416943 GN=mdcB PE=3 SV=1
MM1 pKa = 7.61ALASAGKK8 pKa = 10.05CLLSYY13 pKa = 10.85RR14 pKa = 11.84LDD16 pKa = 3.37EE17 pKa = 4.46SRR19 pKa = 11.84QSRR22 pKa = 11.84SCIDD26 pKa = 3.82SNPSTQEE33 pKa = 3.29EE34 pKa = 4.54SDD36 pKa = 3.77AVHH39 pKa = 6.25CRR41 pKa = 11.84YY42 pKa = 9.48PVDD45 pKa = 3.78HH46 pKa = 7.28KK47 pKa = 11.47SPATGRR53 pKa = 11.84PFSFSCRR60 pKa = 11.84LLVATTAAFPFMLAAGPAYY79 pKa = 10.24ADD81 pKa = 3.69GAGGPNSAALGGSSGINGGAGGGGGVGGVGGANTGANVGAGGNGSATGTGGAGSSGTATGGGSPGGGGSGGAAGTAANPAGVGGGAGGNASCGLCVAGLGGGGGGGGGDD190 pKa = 3.54GFVGATLNGVTGVMTGGAGGNGGNATSNSLLAGADD225 pKa = 3.54GGSGGGGGAGAVLSGSGSTSTAANVSGGAGGSGGTALSSGGAGFGGGGGSGLVDD279 pKa = 3.06SGVNVTNSFSITGGNGGAGGSGAQAGTGRR308 pKa = 11.84SGGNGGAGVTGSSFTLTNTGIVQGGNGGATGGNGSGTGTGVLGAGGVGVQGANLQIVNGGTISGGTAGDD377 pKa = 3.35SSTRR381 pKa = 11.84ANAIAFTGGANSLQLQLGWSFVGNVVSTGGTSTLILGGDD420 pKa = 3.78ATNVSANQAGTSVSTVFALSQIGTVFQGFSGFQKK454 pKa = 9.6TGASTWQLIGSTSDD468 pKa = 3.24STPWTISAGVLQLGNGTTNGSITGNVTNNATLAFDD503 pKa = 4.2PASGTTLTNASEE515 pKa = 4.18ISGTGQVQQIGAGITVLSGVNSYY538 pKa = 11.53AGGTAINAGTLQVAADD554 pKa = 4.06ANLGAAAGTLAFNGGTLNTTASFASSRR581 pKa = 11.84ATTLDD586 pKa = 3.12AGGGTFDD593 pKa = 3.92VGSGTTLTMGGAISGAGLLTKK614 pKa = 9.96TDD616 pKa = 3.43TGSLVLTGTNSYY628 pKa = 11.31AGGTTISAGTLQLGDD643 pKa = 3.48GTTNGSITGNVTNNATLAFDD663 pKa = 4.36PAASTTLTNTSAISGTGQAQQIGSGTTVLTGASTYY698 pKa = 10.47TGGTTISAGVLQLGDD713 pKa = 3.44GTTNGSITGNVTNDD727 pKa = 2.54ATFAFDD733 pKa = 3.8PASGTTMTNPGAISGTGQVQQIGAGTTVLTGVNTYY768 pKa = 9.72TGGTTISGGVLQLGDD783 pKa = 3.57GTTNGSITGSVTNNATLAFDD803 pKa = 4.2PASGTTITNAGVISGSGDD821 pKa = 3.35VQQIGAGITMLSGANTYY838 pKa = 9.63TGGTTIDD845 pKa = 3.38AGTLQVAADD854 pKa = 4.3DD855 pKa = 4.34NLGGAAGALTFNGGTLNTTASFASSRR881 pKa = 11.84ATTLDD886 pKa = 3.12AGGGTFDD893 pKa = 3.92VGSGTTLTMGGTISGAGLLTKK914 pKa = 10.0TDD916 pKa = 3.29TGVLVLTGTNGYY928 pKa = 7.98TGGTTISAGTLQLGNGTTNGSITGNVTNNATLAFDD963 pKa = 4.36PAASTTLTNASAISGTGQVQQIGIGTTVLTGASSYY998 pKa = 10.87TGGTTISAGVLQLGDD1013 pKa = 3.44GTTNGSITGNVTNDD1027 pKa = 2.64ATLAFDD1033 pKa = 4.24PATGTTMTNSGLISGAGQVQQIGAGTTVLSGTNAYY1068 pKa = 8.91TGGTTIDD1075 pKa = 3.58AGTLQLANASAAGTGTVTVDD1095 pKa = 3.36TAAGNTAQGLSLAFSSAGNFANTLSGSGVTTVAAGSALATITGSNASYY1143 pKa = 10.66AGNWQIAGNAAVAPASTTSTANLGTGTVNVAAGGGLTVATSGAFAFNNALTGSGALSASSGGQAFSFGSAAGSAFQGTATLSNDD1227 pKa = 3.03TFALAGANTTALTNATLVLGAGNVTTVGDD1256 pKa = 4.0GNHH1259 pKa = 7.05TIGALTFDD1267 pKa = 3.93GGTAVFDD1274 pKa = 3.82GTAPAQQVATSLVTTGALDD1293 pKa = 3.27ISGSGAVRR1301 pKa = 11.84INVPNPYY1308 pKa = 9.23EE1309 pKa = 4.29PPVPTPSGDD1318 pKa = 3.26LSLFQQSQMNNGLKK1332 pKa = 10.45LIAAASVTGTGSGLALEE1349 pKa = 4.84DD1350 pKa = 3.53QNGNAVSAAQNVNVQQNGNVVAIGTYY1376 pKa = 10.16DD1377 pKa = 3.48YY1378 pKa = 11.84ALGTGASDD1386 pKa = 3.51DD1387 pKa = 3.84GLYY1390 pKa = 10.58VGYY1393 pKa = 10.18QLNQLALQAGQSLTLTEE1410 pKa = 4.98DD1411 pKa = 3.38PGATGADD1418 pKa = 3.35ADD1420 pKa = 4.97LAAQLTGTGNLVIAANDD1437 pKa = 3.55VVSLSNAGNTFTGTTLVQSGTLQANAANVIASSGAVDD1474 pKa = 3.65VEE1476 pKa = 4.8STFDD1480 pKa = 3.44TNGFSQSLNNLTGTAAASIALSGNDD1505 pKa = 3.43TLTLNGGASPNTFAGVIGGAGSLVQASGTEE1535 pKa = 4.13VLTGANTYY1543 pKa = 9.63TGGTTISGGTLQLGDD1558 pKa = 3.6GTTNGSITGNVTNDD1572 pKa = 2.64ATLAFDD1578 pKa = 4.14PAAATTMTNPGLISGSGQVQQMGAGTTVFTGTQTYY1613 pKa = 9.31TGGTTISAGVLQLGDD1628 pKa = 3.57GTTNGAIAGSVTDD1641 pKa = 3.77NATLAFDD1648 pKa = 4.31PATGTTTSHH1657 pKa = 6.65AGVISGLGVVQQIGAGTTVLTGANTYY1683 pKa = 9.63TGGTTISSGVLQLGDD1698 pKa = 3.41GTTNGSIAGSVTNDD1712 pKa = 2.59ATLAFDD1718 pKa = 4.26PAAGTTTTNAGMISGSGDD1736 pKa = 3.31VQQIGAGITVLSGANAYY1753 pKa = 8.8TGGTTLNAGTLQVAADD1769 pKa = 4.3DD1770 pKa = 4.34NLGGAAGALTFNGGTLNTTASFASSRR1796 pKa = 11.84ATTLNAGGGTFGVGSATTLTMGGVISGAGALTKK1829 pKa = 10.1TDD1831 pKa = 3.37AGSLVLTGTNSYY1843 pKa = 9.58TGGTTISVGTLQLGNGTTNGSVAGNVTNNATLAFDD1878 pKa = 4.29PAAGTTMANASVIAGAGQVQQIGTGTTVLTGASSYY1913 pKa = 10.87TGGTTISAGVLQLGDD1928 pKa = 3.57GTTNGAITGNVTDD1941 pKa = 4.06NATLAFDD1948 pKa = 4.4PAASTTLTSAVAISGAGQVQQIGTGTTNLTGDD1980 pKa = 3.24SSTFAGATSVTSGTLLVSGALGDD2003 pKa = 4.14AGSTASVSSGGTLGGGGTLGGSVSVADD2030 pKa = 3.88GTLAPGTTSVSGSPGTLTIGGDD2052 pKa = 3.61LALTGASTLNYY2063 pKa = 10.17HH2064 pKa = 6.9FGQANVVGGPFNDD2077 pKa = 3.5LTEE2080 pKa = 4.31VGGNLTLAGTLNVTSSVGSFDD2101 pKa = 3.39PGIYY2105 pKa = 9.83RR2106 pKa = 11.84IISYY2110 pKa = 10.7GGALTDD2116 pKa = 3.43NGLTVGALPSGTSATVQTSIATEE2139 pKa = 4.06VNLLNTTGVTANFWDD2154 pKa = 4.99GASSPRR2160 pKa = 11.84NNGAVDD2166 pKa = 3.49GGDD2169 pKa = 3.98GVWQSAAGNDD2179 pKa = 2.99NWVASDD2185 pKa = 3.59GALNVPYY2192 pKa = 10.11TSGAFAIFAATPGTVTVDD2210 pKa = 3.19NSAGPVVSGGMQFAIGGYY2228 pKa = 8.42RR2229 pKa = 11.84VEE2231 pKa = 4.82GGPITMAAGGHH2242 pKa = 6.53FIRR2245 pKa = 11.84VGDD2248 pKa = 3.83GTEE2251 pKa = 4.36DD2252 pKa = 3.21GTGYY2256 pKa = 10.78VATIASALTGSGGIDD2271 pKa = 3.43KK2272 pKa = 10.77DD2273 pKa = 4.26DD2274 pKa = 4.84LGTLILTGANSYY2286 pKa = 9.5TGGTTISGGVLQLGDD2301 pKa = 3.57GTTNGSITGNVTNNATLAFDD2321 pKa = 4.24PASGTTMTNPGLISGTGQVQQIGAGTTVFTGTQTYY2356 pKa = 9.35TGGTTIGAGVLQLGDD2371 pKa = 3.51GTTNGALAGNVTNDD2385 pKa = 2.8ATLAFDD2391 pKa = 4.39PAASTTTTNSSVISGTGQVQQIGAGVTVLGGANAYY2426 pKa = 9.0TGGTAINAGTLQVAADD2442 pKa = 3.98NNLGGAAGALTFNGGALNTTASFASSRR2469 pKa = 11.84ATTLNANGGTFDD2481 pKa = 3.62VDD2483 pKa = 3.19AATTLTMSGALSGAGGLTKK2502 pKa = 10.19TDD2504 pKa = 3.02TGTLVLTGTNTYY2516 pKa = 11.07AGGTTISGGTLQLGDD2531 pKa = 3.57GTTNGVITGNVTNNATLAFDD2551 pKa = 4.26PAAGTTLTSAVAISGAGLVQQTGVGTTNLTGDD2583 pKa = 3.34SSAFSGVTNVTGGALLVSGALGGAGSTTTASSGGTLGGGGTLGGNVSVADD2633 pKa = 3.88GTLAPGDD2640 pKa = 3.72ARR2642 pKa = 11.84LGGGPGTLTIGGDD2655 pKa = 3.61LALTGASTLDD2665 pKa = 3.46YY2666 pKa = 11.11HH2667 pKa = 7.19FGQANVIGGPFNDD2680 pKa = 3.89LTDD2683 pKa = 3.41VGGNLTLAGTLNVTSSVGSFDD2704 pKa = 3.39PGIYY2708 pKa = 9.79RR2709 pKa = 11.84IIGYY2713 pKa = 10.06GGGLTNNGLSIGALPAGTTAAVQTSIANEE2742 pKa = 3.95VNLLNTTGITADD2754 pKa = 3.44FWDD2757 pKa = 5.09GASSPRR2763 pKa = 11.84NNGAVDD2769 pKa = 3.49GGDD2772 pKa = 3.98GVWQSAAGNDD2782 pKa = 2.99NWVASDD2788 pKa = 3.59GALNVPYY2795 pKa = 10.28TNGAFAIFAAAPGTVTVDD2813 pKa = 3.17TSVGPVVSGGMQFASDD2829 pKa = 4.39GYY2831 pKa = 9.75RR2832 pKa = 11.84IQGGPITLAAGTDD2845 pKa = 4.51LIRR2848 pKa = 11.84VGDD2851 pKa = 3.99GTEE2854 pKa = 4.22DD2855 pKa = 3.45GAGYY2859 pKa = 10.27VATIASEE2866 pKa = 4.19LTGSGGIDD2874 pKa = 3.43KK2875 pKa = 10.72DD2876 pKa = 4.02DD2877 pKa = 4.88LGTLVLSGANTYY2889 pKa = 9.61TGGTTVSAGTLRR2901 pKa = 11.84LAGGGTLGAAGGATTVSGGLLDD2923 pKa = 5.62LGGTTQTQDD2932 pKa = 2.71AGLTLVSGEE2941 pKa = 4.36VNNGTLFSSTGFALQSGSASASLAGSGALDD2971 pKa = 3.42KK2972 pKa = 8.91TTAGTVTLTGANTYY2986 pKa = 9.38TGATTIDD2993 pKa = 3.74AGTLAIGSGGSIAASSGVTLTAAGAVFDD3021 pKa = 4.11VTAGGNQTVKK3031 pKa = 10.87DD3032 pKa = 3.84LAGVAGSTVDD3042 pKa = 3.74VGASTLNFGTGNSTTFAGSFTGSGALVWQGGGTFSLTGDD3081 pKa = 3.41SSRR3084 pKa = 11.84FGGTTTVAAGTFAIGTDD3101 pKa = 3.57AAPDD3105 pKa = 3.47ASLGGDD3111 pKa = 3.5VVVSGGTLKK3120 pKa = 11.05GFGTIGGNLANRR3132 pKa = 11.84SGTVAPGGSIGTLTVSGNYY3151 pKa = 6.69TQNAGGTLAIEE3162 pKa = 4.39VNPASASQLKK3172 pKa = 10.6VGGAASLDD3180 pKa = 3.55GKK3182 pKa = 10.5LALLFAPGTYY3192 pKa = 10.58NPTSYY3197 pKa = 11.42KK3198 pKa = 10.63LLTAQSVGGTFSAVSGSNPGGLTQSIDD3225 pKa = 3.45YY3226 pKa = 10.44HH3227 pKa = 6.2PADD3230 pKa = 3.24VTLRR3234 pKa = 11.84LSTPSPSSPSLPPPPLVVAPDD3255 pKa = 3.32NDD3257 pKa = 4.41TIYY3260 pKa = 10.31TALTSIAVLNAQQINGILLDD3280 pKa = 4.34RR3281 pKa = 11.84IGQRR3285 pKa = 11.84RR3286 pKa = 11.84AGIADD3291 pKa = 3.75GPVAGLGDD3299 pKa = 4.3PAPQLQYY3306 pKa = 11.53ASAFGPLPAVQSLPTAARR3324 pKa = 11.84GAWFRR3329 pKa = 11.84GIGNFTSLSGRR3340 pKa = 11.84AGAPGFTGSTGGFLLGYY3357 pKa = 8.72DD3358 pKa = 4.27QPFADD3363 pKa = 3.59NAYY3366 pKa = 9.92LGVAGGYY3373 pKa = 10.17LNSSVDD3379 pKa = 3.49EE3380 pKa = 4.74HH3381 pKa = 6.25STSSGSIEE3389 pKa = 3.99SARR3392 pKa = 11.84ISLYY3396 pKa = 10.5GGVVLGPSLFTATAGYY3412 pKa = 9.93AHH3414 pKa = 7.47DD3415 pKa = 3.49WMRR3418 pKa = 11.84TEE3420 pKa = 4.85RR3421 pKa = 11.84GIAGIGTARR3430 pKa = 11.84EE3431 pKa = 3.98SHH3433 pKa = 6.39GGDD3436 pKa = 2.97EE3437 pKa = 4.48ATFAGQWSMPLAIPGLAGGSATLTPKK3463 pKa = 10.7VGIQYY3468 pKa = 9.53VHH3470 pKa = 6.99LSEE3473 pKa = 6.05GGFTDD3478 pKa = 3.91TGAGGFDD3485 pKa = 3.92LGADD3489 pKa = 3.54SRR3491 pKa = 11.84SADD3494 pKa = 3.02SFQPYY3499 pKa = 9.98FGFALGQRR3507 pKa = 11.84FTTRR3511 pKa = 11.84GGTDD3515 pKa = 2.94ITPEE3519 pKa = 3.87LRR3521 pKa = 11.84VGYY3524 pKa = 10.21AYY3526 pKa = 7.78EE3527 pKa = 4.04TLSNARR3533 pKa = 11.84RR3534 pKa = 11.84MTVIAADD3541 pKa = 3.8GTAFPVTGVPPSRR3554 pKa = 11.84NQLTAGIGVTMSAGPNLSFYY3574 pKa = 11.33ANYY3577 pKa = 10.41DD3578 pKa = 3.68AILPVANTRR3587 pKa = 11.84SQTFQAGLRR3596 pKa = 11.84WRR3598 pKa = 11.84FF3599 pKa = 3.12
MM1 pKa = 7.61ALASAGKK8 pKa = 10.05CLLSYY13 pKa = 10.85RR14 pKa = 11.84LDD16 pKa = 3.37EE17 pKa = 4.46SRR19 pKa = 11.84QSRR22 pKa = 11.84SCIDD26 pKa = 3.82SNPSTQEE33 pKa = 3.29EE34 pKa = 4.54SDD36 pKa = 3.77AVHH39 pKa = 6.25CRR41 pKa = 11.84YY42 pKa = 9.48PVDD45 pKa = 3.78HH46 pKa = 7.28KK47 pKa = 11.47SPATGRR53 pKa = 11.84PFSFSCRR60 pKa = 11.84LLVATTAAFPFMLAAGPAYY79 pKa = 10.24ADD81 pKa = 3.69GAGGPNSAALGGSSGINGGAGGGGGVGGVGGANTGANVGAGGNGSATGTGGAGSSGTATGGGSPGGGGSGGAAGTAANPAGVGGGAGGNASCGLCVAGLGGGGGGGGGDD190 pKa = 3.54GFVGATLNGVTGVMTGGAGGNGGNATSNSLLAGADD225 pKa = 3.54GGSGGGGGAGAVLSGSGSTSTAANVSGGAGGSGGTALSSGGAGFGGGGGSGLVDD279 pKa = 3.06SGVNVTNSFSITGGNGGAGGSGAQAGTGRR308 pKa = 11.84SGGNGGAGVTGSSFTLTNTGIVQGGNGGATGGNGSGTGTGVLGAGGVGVQGANLQIVNGGTISGGTAGDD377 pKa = 3.35SSTRR381 pKa = 11.84ANAIAFTGGANSLQLQLGWSFVGNVVSTGGTSTLILGGDD420 pKa = 3.78ATNVSANQAGTSVSTVFALSQIGTVFQGFSGFQKK454 pKa = 9.6TGASTWQLIGSTSDD468 pKa = 3.24STPWTISAGVLQLGNGTTNGSITGNVTNNATLAFDD503 pKa = 4.2PASGTTLTNASEE515 pKa = 4.18ISGTGQVQQIGAGITVLSGVNSYY538 pKa = 11.53AGGTAINAGTLQVAADD554 pKa = 4.06ANLGAAAGTLAFNGGTLNTTASFASSRR581 pKa = 11.84ATTLDD586 pKa = 3.12AGGGTFDD593 pKa = 3.92VGSGTTLTMGGAISGAGLLTKK614 pKa = 9.96TDD616 pKa = 3.43TGSLVLTGTNSYY628 pKa = 11.31AGGTTISAGTLQLGDD643 pKa = 3.48GTTNGSITGNVTNNATLAFDD663 pKa = 4.36PAASTTLTNTSAISGTGQAQQIGSGTTVLTGASTYY698 pKa = 10.47TGGTTISAGVLQLGDD713 pKa = 3.44GTTNGSITGNVTNDD727 pKa = 2.54ATFAFDD733 pKa = 3.8PASGTTMTNPGAISGTGQVQQIGAGTTVLTGVNTYY768 pKa = 9.72TGGTTISGGVLQLGDD783 pKa = 3.57GTTNGSITGSVTNNATLAFDD803 pKa = 4.2PASGTTITNAGVISGSGDD821 pKa = 3.35VQQIGAGITMLSGANTYY838 pKa = 9.63TGGTTIDD845 pKa = 3.38AGTLQVAADD854 pKa = 4.3DD855 pKa = 4.34NLGGAAGALTFNGGTLNTTASFASSRR881 pKa = 11.84ATTLDD886 pKa = 3.12AGGGTFDD893 pKa = 3.92VGSGTTLTMGGTISGAGLLTKK914 pKa = 10.0TDD916 pKa = 3.29TGVLVLTGTNGYY928 pKa = 7.98TGGTTISAGTLQLGNGTTNGSITGNVTNNATLAFDD963 pKa = 4.36PAASTTLTNASAISGTGQVQQIGIGTTVLTGASSYY998 pKa = 10.87TGGTTISAGVLQLGDD1013 pKa = 3.44GTTNGSITGNVTNDD1027 pKa = 2.64ATLAFDD1033 pKa = 4.24PATGTTMTNSGLISGAGQVQQIGAGTTVLSGTNAYY1068 pKa = 8.91TGGTTIDD1075 pKa = 3.58AGTLQLANASAAGTGTVTVDD1095 pKa = 3.36TAAGNTAQGLSLAFSSAGNFANTLSGSGVTTVAAGSALATITGSNASYY1143 pKa = 10.66AGNWQIAGNAAVAPASTTSTANLGTGTVNVAAGGGLTVATSGAFAFNNALTGSGALSASSGGQAFSFGSAAGSAFQGTATLSNDD1227 pKa = 3.03TFALAGANTTALTNATLVLGAGNVTTVGDD1256 pKa = 4.0GNHH1259 pKa = 7.05TIGALTFDD1267 pKa = 3.93GGTAVFDD1274 pKa = 3.82GTAPAQQVATSLVTTGALDD1293 pKa = 3.27ISGSGAVRR1301 pKa = 11.84INVPNPYY1308 pKa = 9.23EE1309 pKa = 4.29PPVPTPSGDD1318 pKa = 3.26LSLFQQSQMNNGLKK1332 pKa = 10.45LIAAASVTGTGSGLALEE1349 pKa = 4.84DD1350 pKa = 3.53QNGNAVSAAQNVNVQQNGNVVAIGTYY1376 pKa = 10.16DD1377 pKa = 3.48YY1378 pKa = 11.84ALGTGASDD1386 pKa = 3.51DD1387 pKa = 3.84GLYY1390 pKa = 10.58VGYY1393 pKa = 10.18QLNQLALQAGQSLTLTEE1410 pKa = 4.98DD1411 pKa = 3.38PGATGADD1418 pKa = 3.35ADD1420 pKa = 4.97LAAQLTGTGNLVIAANDD1437 pKa = 3.55VVSLSNAGNTFTGTTLVQSGTLQANAANVIASSGAVDD1474 pKa = 3.65VEE1476 pKa = 4.8STFDD1480 pKa = 3.44TNGFSQSLNNLTGTAAASIALSGNDD1505 pKa = 3.43TLTLNGGASPNTFAGVIGGAGSLVQASGTEE1535 pKa = 4.13VLTGANTYY1543 pKa = 9.63TGGTTISGGTLQLGDD1558 pKa = 3.6GTTNGSITGNVTNDD1572 pKa = 2.64ATLAFDD1578 pKa = 4.14PAAATTMTNPGLISGSGQVQQMGAGTTVFTGTQTYY1613 pKa = 9.31TGGTTISAGVLQLGDD1628 pKa = 3.57GTTNGAIAGSVTDD1641 pKa = 3.77NATLAFDD1648 pKa = 4.31PATGTTTSHH1657 pKa = 6.65AGVISGLGVVQQIGAGTTVLTGANTYY1683 pKa = 9.63TGGTTISSGVLQLGDD1698 pKa = 3.41GTTNGSIAGSVTNDD1712 pKa = 2.59ATLAFDD1718 pKa = 4.26PAAGTTTTNAGMISGSGDD1736 pKa = 3.31VQQIGAGITVLSGANAYY1753 pKa = 8.8TGGTTLNAGTLQVAADD1769 pKa = 4.3DD1770 pKa = 4.34NLGGAAGALTFNGGTLNTTASFASSRR1796 pKa = 11.84ATTLNAGGGTFGVGSATTLTMGGVISGAGALTKK1829 pKa = 10.1TDD1831 pKa = 3.37AGSLVLTGTNSYY1843 pKa = 9.58TGGTTISVGTLQLGNGTTNGSVAGNVTNNATLAFDD1878 pKa = 4.29PAAGTTMANASVIAGAGQVQQIGTGTTVLTGASSYY1913 pKa = 10.87TGGTTISAGVLQLGDD1928 pKa = 3.57GTTNGAITGNVTDD1941 pKa = 4.06NATLAFDD1948 pKa = 4.4PAASTTLTSAVAISGAGQVQQIGTGTTNLTGDD1980 pKa = 3.24SSTFAGATSVTSGTLLVSGALGDD2003 pKa = 4.14AGSTASVSSGGTLGGGGTLGGSVSVADD2030 pKa = 3.88GTLAPGTTSVSGSPGTLTIGGDD2052 pKa = 3.61LALTGASTLNYY2063 pKa = 10.17HH2064 pKa = 6.9FGQANVVGGPFNDD2077 pKa = 3.5LTEE2080 pKa = 4.31VGGNLTLAGTLNVTSSVGSFDD2101 pKa = 3.39PGIYY2105 pKa = 9.83RR2106 pKa = 11.84IISYY2110 pKa = 10.7GGALTDD2116 pKa = 3.43NGLTVGALPSGTSATVQTSIATEE2139 pKa = 4.06VNLLNTTGVTANFWDD2154 pKa = 4.99GASSPRR2160 pKa = 11.84NNGAVDD2166 pKa = 3.49GGDD2169 pKa = 3.98GVWQSAAGNDD2179 pKa = 2.99NWVASDD2185 pKa = 3.59GALNVPYY2192 pKa = 10.11TSGAFAIFAATPGTVTVDD2210 pKa = 3.19NSAGPVVSGGMQFAIGGYY2228 pKa = 8.42RR2229 pKa = 11.84VEE2231 pKa = 4.82GGPITMAAGGHH2242 pKa = 6.53FIRR2245 pKa = 11.84VGDD2248 pKa = 3.83GTEE2251 pKa = 4.36DD2252 pKa = 3.21GTGYY2256 pKa = 10.78VATIASALTGSGGIDD2271 pKa = 3.43KK2272 pKa = 10.77DD2273 pKa = 4.26DD2274 pKa = 4.84LGTLILTGANSYY2286 pKa = 9.5TGGTTISGGVLQLGDD2301 pKa = 3.57GTTNGSITGNVTNNATLAFDD2321 pKa = 4.24PASGTTMTNPGLISGTGQVQQIGAGTTVFTGTQTYY2356 pKa = 9.35TGGTTIGAGVLQLGDD2371 pKa = 3.51GTTNGALAGNVTNDD2385 pKa = 2.8ATLAFDD2391 pKa = 4.39PAASTTTTNSSVISGTGQVQQIGAGVTVLGGANAYY2426 pKa = 9.0TGGTAINAGTLQVAADD2442 pKa = 3.98NNLGGAAGALTFNGGALNTTASFASSRR2469 pKa = 11.84ATTLNANGGTFDD2481 pKa = 3.62VDD2483 pKa = 3.19AATTLTMSGALSGAGGLTKK2502 pKa = 10.19TDD2504 pKa = 3.02TGTLVLTGTNTYY2516 pKa = 11.07AGGTTISGGTLQLGDD2531 pKa = 3.57GTTNGVITGNVTNNATLAFDD2551 pKa = 4.26PAAGTTLTSAVAISGAGLVQQTGVGTTNLTGDD2583 pKa = 3.34SSAFSGVTNVTGGALLVSGALGGAGSTTTASSGGTLGGGGTLGGNVSVADD2633 pKa = 3.88GTLAPGDD2640 pKa = 3.72ARR2642 pKa = 11.84LGGGPGTLTIGGDD2655 pKa = 3.61LALTGASTLDD2665 pKa = 3.46YY2666 pKa = 11.11HH2667 pKa = 7.19FGQANVIGGPFNDD2680 pKa = 3.89LTDD2683 pKa = 3.41VGGNLTLAGTLNVTSSVGSFDD2704 pKa = 3.39PGIYY2708 pKa = 9.79RR2709 pKa = 11.84IIGYY2713 pKa = 10.06GGGLTNNGLSIGALPAGTTAAVQTSIANEE2742 pKa = 3.95VNLLNTTGITADD2754 pKa = 3.44FWDD2757 pKa = 5.09GASSPRR2763 pKa = 11.84NNGAVDD2769 pKa = 3.49GGDD2772 pKa = 3.98GVWQSAAGNDD2782 pKa = 2.99NWVASDD2788 pKa = 3.59GALNVPYY2795 pKa = 10.28TNGAFAIFAAAPGTVTVDD2813 pKa = 3.17TSVGPVVSGGMQFASDD2829 pKa = 4.39GYY2831 pKa = 9.75RR2832 pKa = 11.84IQGGPITLAAGTDD2845 pKa = 4.51LIRR2848 pKa = 11.84VGDD2851 pKa = 3.99GTEE2854 pKa = 4.22DD2855 pKa = 3.45GAGYY2859 pKa = 10.27VATIASEE2866 pKa = 4.19LTGSGGIDD2874 pKa = 3.43KK2875 pKa = 10.72DD2876 pKa = 4.02DD2877 pKa = 4.88LGTLVLSGANTYY2889 pKa = 9.61TGGTTVSAGTLRR2901 pKa = 11.84LAGGGTLGAAGGATTVSGGLLDD2923 pKa = 5.62LGGTTQTQDD2932 pKa = 2.71AGLTLVSGEE2941 pKa = 4.36VNNGTLFSSTGFALQSGSASASLAGSGALDD2971 pKa = 3.42KK2972 pKa = 8.91TTAGTVTLTGANTYY2986 pKa = 9.38TGATTIDD2993 pKa = 3.74AGTLAIGSGGSIAASSGVTLTAAGAVFDD3021 pKa = 4.11VTAGGNQTVKK3031 pKa = 10.87DD3032 pKa = 3.84LAGVAGSTVDD3042 pKa = 3.74VGASTLNFGTGNSTTFAGSFTGSGALVWQGGGTFSLTGDD3081 pKa = 3.41SSRR3084 pKa = 11.84FGGTTTVAAGTFAIGTDD3101 pKa = 3.57AAPDD3105 pKa = 3.47ASLGGDD3111 pKa = 3.5VVVSGGTLKK3120 pKa = 11.05GFGTIGGNLANRR3132 pKa = 11.84SGTVAPGGSIGTLTVSGNYY3151 pKa = 6.69TQNAGGTLAIEE3162 pKa = 4.39VNPASASQLKK3172 pKa = 10.6VGGAASLDD3180 pKa = 3.55GKK3182 pKa = 10.5LALLFAPGTYY3192 pKa = 10.58NPTSYY3197 pKa = 11.42KK3198 pKa = 10.63LLTAQSVGGTFSAVSGSNPGGLTQSIDD3225 pKa = 3.45YY3226 pKa = 10.44HH3227 pKa = 6.2PADD3230 pKa = 3.24VTLRR3234 pKa = 11.84LSTPSPSSPSLPPPPLVVAPDD3255 pKa = 3.32NDD3257 pKa = 4.41TIYY3260 pKa = 10.31TALTSIAVLNAQQINGILLDD3280 pKa = 4.34RR3281 pKa = 11.84IGQRR3285 pKa = 11.84RR3286 pKa = 11.84AGIADD3291 pKa = 3.75GPVAGLGDD3299 pKa = 4.3PAPQLQYY3306 pKa = 11.53ASAFGPLPAVQSLPTAARR3324 pKa = 11.84GAWFRR3329 pKa = 11.84GIGNFTSLSGRR3340 pKa = 11.84AGAPGFTGSTGGFLLGYY3357 pKa = 8.72DD3358 pKa = 4.27QPFADD3363 pKa = 3.59NAYY3366 pKa = 9.92LGVAGGYY3373 pKa = 10.17LNSSVDD3379 pKa = 3.49EE3380 pKa = 4.74HH3381 pKa = 6.25STSSGSIEE3389 pKa = 3.99SARR3392 pKa = 11.84ISLYY3396 pKa = 10.5GGVVLGPSLFTATAGYY3412 pKa = 9.93AHH3414 pKa = 7.47DD3415 pKa = 3.49WMRR3418 pKa = 11.84TEE3420 pKa = 4.85RR3421 pKa = 11.84GIAGIGTARR3430 pKa = 11.84EE3431 pKa = 3.98SHH3433 pKa = 6.39GGDD3436 pKa = 2.97EE3437 pKa = 4.48ATFAGQWSMPLAIPGLAGGSATLTPKK3463 pKa = 10.7VGIQYY3468 pKa = 9.53VHH3470 pKa = 6.99LSEE3473 pKa = 6.05GGFTDD3478 pKa = 3.91TGAGGFDD3485 pKa = 3.92LGADD3489 pKa = 3.54SRR3491 pKa = 11.84SADD3494 pKa = 3.02SFQPYY3499 pKa = 9.98FGFALGQRR3507 pKa = 11.84FTTRR3511 pKa = 11.84GGTDD3515 pKa = 2.94ITPEE3519 pKa = 3.87LRR3521 pKa = 11.84VGYY3524 pKa = 10.21AYY3526 pKa = 7.78EE3527 pKa = 4.04TLSNARR3533 pKa = 11.84RR3534 pKa = 11.84MTVIAADD3541 pKa = 3.8GTAFPVTGVPPSRR3554 pKa = 11.84NQLTAGIGVTMSAGPNLSFYY3574 pKa = 11.33ANYY3577 pKa = 10.41DD3578 pKa = 3.68AILPVANTRR3587 pKa = 11.84SQTFQAGLRR3596 pKa = 11.84WRR3598 pKa = 11.84FF3599 pKa = 3.12
Molecular weight: 340.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7H559|A0A1H7H559_9BURK Chromosome segregation DNA-binding protein OS=Paraburkholderia caballeronis OX=416943 GN=SAMN05192542_10268 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LAII44 pKa = 4.0
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.65GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LAII44 pKa = 4.0
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2028450 |
29 |
6079 |
327.8 |
35.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.413 ± 0.044 | 0.898 ± 0.01 |
5.811 ± 0.028 | 4.813 ± 0.037 |
3.621 ± 0.02 | 8.429 ± 0.046 |
2.292 ± 0.016 | 4.424 ± 0.023 |
2.619 ± 0.027 | 10.191 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.177 ± 0.015 | 2.681 ± 0.024 |
5.279 ± 0.024 | 3.5 ± 0.017 |
7.38 ± 0.045 | 5.449 ± 0.029 |
5.408 ± 0.041 | 7.859 ± 0.028 |
1.371 ± 0.012 | 2.386 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
