
Sinobaca qinghaiensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Sporolactobacillaceae; Sinobaca
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
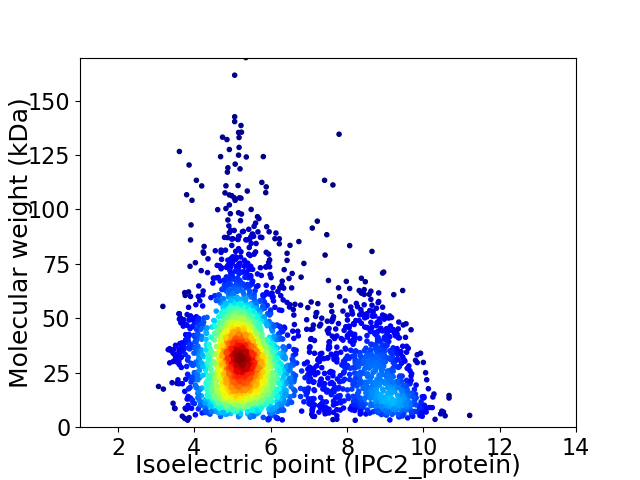
Virtual 2D-PAGE plot for 3379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A419V5Q1|A0A419V5Q1_9BACL Protein RecA OS=Sinobaca qinghaiensis OX=342944 GN=recA PE=3 SV=1
MM1 pKa = 7.37NKK3 pKa = 9.74LMQFGFISVIVISLTACSEE22 pKa = 4.14EE23 pKa = 4.22SSKK26 pKa = 10.77EE27 pKa = 3.63IPKK30 pKa = 10.51AKK32 pKa = 10.02SAEE35 pKa = 4.18TSEE38 pKa = 4.65DD39 pKa = 3.33PQGFSEE45 pKa = 4.4SSKK48 pKa = 10.72EE49 pKa = 3.78GDD51 pKa = 2.99IVTVEE56 pKa = 4.0GQITYY61 pKa = 10.77AEE63 pKa = 5.02GGTAPGEE70 pKa = 4.26LYY72 pKa = 11.0GDD74 pKa = 3.66TQYY77 pKa = 11.47TIITDD82 pKa = 3.75ANGEE86 pKa = 3.89EE87 pKa = 5.08AYY89 pKa = 10.52DD90 pKa = 3.57ITHH93 pKa = 6.54SANEE97 pKa = 4.02TLMIGDD103 pKa = 4.0DD104 pKa = 3.69VKK106 pKa = 11.07VTGEE110 pKa = 3.92YY111 pKa = 10.65LGVNTGMEE119 pKa = 4.09IPAIDD124 pKa = 3.94GSKK127 pKa = 10.65VEE129 pKa = 4.44VINYY133 pKa = 9.18NNGVVTIYY141 pKa = 11.13DD142 pKa = 3.57EE143 pKa = 5.17DD144 pKa = 4.43LEE146 pKa = 5.0SNADD150 pKa = 3.48YY151 pKa = 11.29SKK153 pKa = 11.2DD154 pKa = 3.0ITIEE158 pKa = 3.85NEE160 pKa = 3.92VFFNTEE166 pKa = 3.21AEE168 pKa = 4.24FFVDD172 pKa = 3.86SDD174 pKa = 3.58EE175 pKa = 4.35TVYY178 pKa = 10.44FAEE181 pKa = 4.58LKK183 pKa = 10.99NNGSEE188 pKa = 4.09PLDD191 pKa = 3.48ISSVSITWYY200 pKa = 10.53DD201 pKa = 3.24NDD203 pKa = 5.11DD204 pKa = 3.94NVSMVTDD211 pKa = 3.41SSIEE215 pKa = 3.86VFPTVIEE222 pKa = 4.45PGEE225 pKa = 4.09KK226 pKa = 10.45GYY228 pKa = 10.49ISYY231 pKa = 11.22SDD233 pKa = 3.39VTSGVKK239 pKa = 9.87DD240 pKa = 3.75LKK242 pKa = 10.22NTEE245 pKa = 4.54LNLTPYY251 pKa = 10.73ASYY254 pKa = 11.37SSTEE258 pKa = 3.91EE259 pKa = 3.75LKK261 pKa = 10.94VEE263 pKa = 4.11NDD265 pKa = 3.02HH266 pKa = 7.34FEE268 pKa = 4.38AKK270 pKa = 9.16TDD272 pKa = 3.39SWDD275 pKa = 3.15EE276 pKa = 4.49SYY278 pKa = 11.61VHH280 pKa = 6.79ILGEE284 pKa = 3.95ITNQSEE290 pKa = 4.5TPLYY294 pKa = 9.2NHH296 pKa = 6.32TVYY299 pKa = 11.03SVFRR303 pKa = 11.84DD304 pKa = 3.51EE305 pKa = 4.88NGNFLGLDD313 pKa = 3.64STSGDD318 pKa = 3.1TTLQPNQTSGFEE330 pKa = 3.99ISGEE334 pKa = 4.08DD335 pKa = 3.5LPKK338 pKa = 10.72EE339 pKa = 3.99QIEE342 pKa = 4.72KK343 pKa = 9.93IDD345 pKa = 3.68SVEE348 pKa = 3.79NSAYY352 pKa = 10.59ADD354 pKa = 3.59TFDD357 pKa = 3.79YY358 pKa = 11.41
MM1 pKa = 7.37NKK3 pKa = 9.74LMQFGFISVIVISLTACSEE22 pKa = 4.14EE23 pKa = 4.22SSKK26 pKa = 10.77EE27 pKa = 3.63IPKK30 pKa = 10.51AKK32 pKa = 10.02SAEE35 pKa = 4.18TSEE38 pKa = 4.65DD39 pKa = 3.33PQGFSEE45 pKa = 4.4SSKK48 pKa = 10.72EE49 pKa = 3.78GDD51 pKa = 2.99IVTVEE56 pKa = 4.0GQITYY61 pKa = 10.77AEE63 pKa = 5.02GGTAPGEE70 pKa = 4.26LYY72 pKa = 11.0GDD74 pKa = 3.66TQYY77 pKa = 11.47TIITDD82 pKa = 3.75ANGEE86 pKa = 3.89EE87 pKa = 5.08AYY89 pKa = 10.52DD90 pKa = 3.57ITHH93 pKa = 6.54SANEE97 pKa = 4.02TLMIGDD103 pKa = 4.0DD104 pKa = 3.69VKK106 pKa = 11.07VTGEE110 pKa = 3.92YY111 pKa = 10.65LGVNTGMEE119 pKa = 4.09IPAIDD124 pKa = 3.94GSKK127 pKa = 10.65VEE129 pKa = 4.44VINYY133 pKa = 9.18NNGVVTIYY141 pKa = 11.13DD142 pKa = 3.57EE143 pKa = 5.17DD144 pKa = 4.43LEE146 pKa = 5.0SNADD150 pKa = 3.48YY151 pKa = 11.29SKK153 pKa = 11.2DD154 pKa = 3.0ITIEE158 pKa = 3.85NEE160 pKa = 3.92VFFNTEE166 pKa = 3.21AEE168 pKa = 4.24FFVDD172 pKa = 3.86SDD174 pKa = 3.58EE175 pKa = 4.35TVYY178 pKa = 10.44FAEE181 pKa = 4.58LKK183 pKa = 10.99NNGSEE188 pKa = 4.09PLDD191 pKa = 3.48ISSVSITWYY200 pKa = 10.53DD201 pKa = 3.24NDD203 pKa = 5.11DD204 pKa = 3.94NVSMVTDD211 pKa = 3.41SSIEE215 pKa = 3.86VFPTVIEE222 pKa = 4.45PGEE225 pKa = 4.09KK226 pKa = 10.45GYY228 pKa = 10.49ISYY231 pKa = 11.22SDD233 pKa = 3.39VTSGVKK239 pKa = 9.87DD240 pKa = 3.75LKK242 pKa = 10.22NTEE245 pKa = 4.54LNLTPYY251 pKa = 10.73ASYY254 pKa = 11.37SSTEE258 pKa = 3.91EE259 pKa = 3.75LKK261 pKa = 10.94VEE263 pKa = 4.11NDD265 pKa = 3.02HH266 pKa = 7.34FEE268 pKa = 4.38AKK270 pKa = 9.16TDD272 pKa = 3.39SWDD275 pKa = 3.15EE276 pKa = 4.49SYY278 pKa = 11.61VHH280 pKa = 6.79ILGEE284 pKa = 3.95ITNQSEE290 pKa = 4.5TPLYY294 pKa = 9.2NHH296 pKa = 6.32TVYY299 pKa = 11.03SVFRR303 pKa = 11.84DD304 pKa = 3.51EE305 pKa = 4.88NGNFLGLDD313 pKa = 3.64STSGDD318 pKa = 3.1TTLQPNQTSGFEE330 pKa = 3.99ISGEE334 pKa = 4.08DD335 pKa = 3.5LPKK338 pKa = 10.72EE339 pKa = 3.99QIEE342 pKa = 4.72KK343 pKa = 9.93IDD345 pKa = 3.68SVEE348 pKa = 3.79NSAYY352 pKa = 10.59ADD354 pKa = 3.59TFDD357 pKa = 3.79YY358 pKa = 11.41
Molecular weight: 39.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A419UVY3|A0A419UVY3_9BACL Fructokinase OS=Sinobaca qinghaiensis OX=342944 GN=ATL39_3232 PE=3 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.21KK15 pKa = 8.76NHH17 pKa = 4.93GFRR20 pKa = 11.84TRR22 pKa = 11.84MSTKK26 pKa = 9.81NGRR29 pKa = 11.84HH30 pKa = 6.38IIRR33 pKa = 11.84NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.63GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.21KK15 pKa = 8.76NHH17 pKa = 4.93GFRR20 pKa = 11.84TRR22 pKa = 11.84MSTKK26 pKa = 9.81NGRR29 pKa = 11.84HH30 pKa = 6.38IIRR33 pKa = 11.84NRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.63GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996966 |
29 |
1530 |
295.0 |
32.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.201 ± 0.051 | 0.647 ± 0.012 |
5.151 ± 0.035 | 8.273 ± 0.06 |
4.316 ± 0.037 | 7.15 ± 0.044 |
2.176 ± 0.022 | 7.087 ± 0.039 |
5.991 ± 0.045 | 9.42 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.974 ± 0.021 | 3.777 ± 0.028 |
3.802 ± 0.022 | 3.718 ± 0.031 |
4.293 ± 0.032 | 6.243 ± 0.03 |
5.502 ± 0.029 | 6.864 ± 0.037 |
1.077 ± 0.016 | 3.338 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
