
Hubei tombus-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
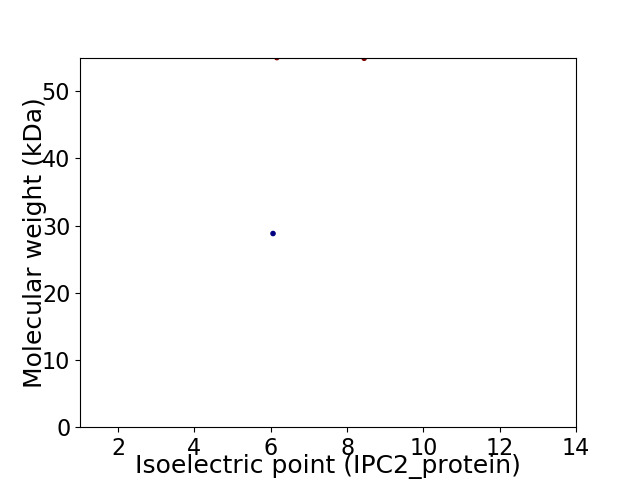
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGV0|A0A1L3KGV0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 11 OX=1923257 PE=4 SV=1
MM1 pKa = 8.11EE2 pKa = 4.18YY3 pKa = 9.98TLNVPTRR10 pKa = 11.84GPLGGSSRR18 pKa = 11.84QVNSQYY24 pKa = 11.16SSRR27 pKa = 11.84SYY29 pKa = 10.58GYY31 pKa = 9.21YY32 pKa = 9.31RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.61AASRR39 pKa = 11.84HH40 pKa = 4.85FKK42 pKa = 10.53PIGEE46 pKa = 5.03AFDD49 pKa = 4.64EE50 pKa = 4.27MDD52 pKa = 3.75GQNIPTQHH60 pKa = 6.54LRR62 pKa = 11.84DD63 pKa = 3.54NMEE66 pKa = 4.34RR67 pKa = 11.84LDD69 pKa = 4.45AEE71 pKa = 4.44PSVQEE76 pKa = 4.16SMIAGYY82 pKa = 10.17LLPTPEE88 pKa = 4.24TFQNGHH94 pKa = 6.37IPAHH98 pKa = 6.88DD99 pKa = 4.49DD100 pKa = 3.37EE101 pKa = 5.17PLQHH105 pKa = 6.88HH106 pKa = 6.27MLEE109 pKa = 4.64KK110 pKa = 10.41FDD112 pKa = 5.47IIPGYY117 pKa = 10.67DD118 pKa = 3.09VTLNEE123 pKa = 4.41QPVEE127 pKa = 4.11NQQVMVCTEE136 pKa = 4.98LLRR139 pKa = 11.84QLQTYY144 pKa = 10.11AAFKK148 pKa = 10.37PRR150 pKa = 11.84TSEE153 pKa = 4.58LLLSLKK159 pKa = 10.86NKK161 pKa = 9.66GSQLARR167 pKa = 11.84PYY169 pKa = 11.21ANSAEE174 pKa = 4.23WKK176 pKa = 8.13HH177 pKa = 5.13QQVLMCSAIAFEE189 pKa = 4.82PSTEE193 pKa = 4.2EE194 pKa = 3.54ITALAHH200 pKa = 6.21ISSSAVATKK209 pKa = 8.28WTFVNDD215 pKa = 3.57RR216 pKa = 11.84LAGGNKK222 pKa = 9.84DD223 pKa = 5.29LIDD226 pKa = 4.08CRR228 pKa = 11.84VLKK231 pKa = 10.49PKK233 pKa = 10.38LATRR237 pKa = 11.84FLSWIYY243 pKa = 10.54PSVTKK248 pKa = 7.92QTPVPVNN255 pKa = 3.27
MM1 pKa = 8.11EE2 pKa = 4.18YY3 pKa = 9.98TLNVPTRR10 pKa = 11.84GPLGGSSRR18 pKa = 11.84QVNSQYY24 pKa = 11.16SSRR27 pKa = 11.84SYY29 pKa = 10.58GYY31 pKa = 9.21YY32 pKa = 9.31RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.61AASRR39 pKa = 11.84HH40 pKa = 4.85FKK42 pKa = 10.53PIGEE46 pKa = 5.03AFDD49 pKa = 4.64EE50 pKa = 4.27MDD52 pKa = 3.75GQNIPTQHH60 pKa = 6.54LRR62 pKa = 11.84DD63 pKa = 3.54NMEE66 pKa = 4.34RR67 pKa = 11.84LDD69 pKa = 4.45AEE71 pKa = 4.44PSVQEE76 pKa = 4.16SMIAGYY82 pKa = 10.17LLPTPEE88 pKa = 4.24TFQNGHH94 pKa = 6.37IPAHH98 pKa = 6.88DD99 pKa = 4.49DD100 pKa = 3.37EE101 pKa = 5.17PLQHH105 pKa = 6.88HH106 pKa = 6.27MLEE109 pKa = 4.64KK110 pKa = 10.41FDD112 pKa = 5.47IIPGYY117 pKa = 10.67DD118 pKa = 3.09VTLNEE123 pKa = 4.41QPVEE127 pKa = 4.11NQQVMVCTEE136 pKa = 4.98LLRR139 pKa = 11.84QLQTYY144 pKa = 10.11AAFKK148 pKa = 10.37PRR150 pKa = 11.84TSEE153 pKa = 4.58LLLSLKK159 pKa = 10.86NKK161 pKa = 9.66GSQLARR167 pKa = 11.84PYY169 pKa = 11.21ANSAEE174 pKa = 4.23WKK176 pKa = 8.13HH177 pKa = 5.13QQVLMCSAIAFEE189 pKa = 4.82PSTEE193 pKa = 4.2EE194 pKa = 3.54ITALAHH200 pKa = 6.21ISSSAVATKK209 pKa = 8.28WTFVNDD215 pKa = 3.57RR216 pKa = 11.84LAGGNKK222 pKa = 9.84DD223 pKa = 5.29LIDD226 pKa = 4.08CRR228 pKa = 11.84VLKK231 pKa = 10.49PKK233 pKa = 10.38LATRR237 pKa = 11.84FLSWIYY243 pKa = 10.54PSVTKK248 pKa = 7.92QTPVPVNN255 pKa = 3.27
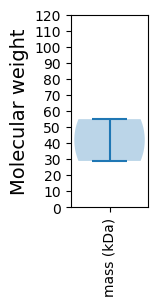
Molecular weight: 28.81 kDa
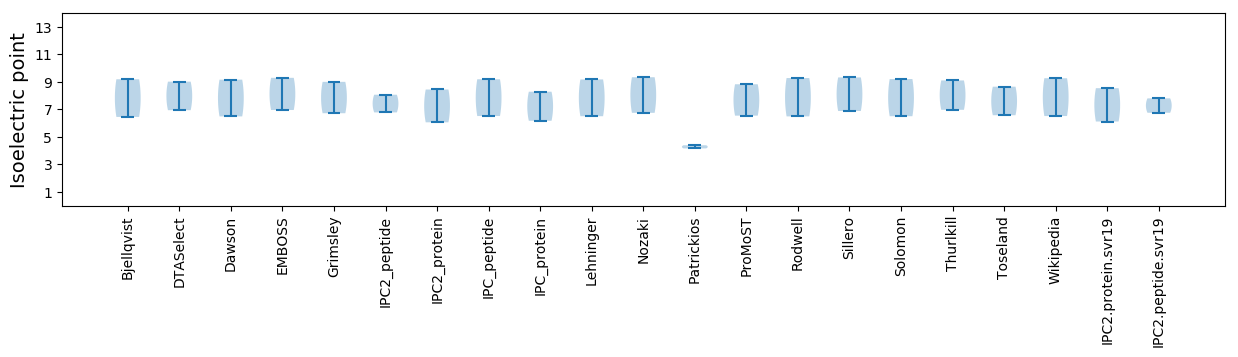
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGV0|A0A1L3KGV0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 11 OX=1923257 PE=4 SV=1
MM1 pKa = 7.67SEE3 pKa = 3.95RR4 pKa = 11.84DD5 pKa = 3.42KK6 pKa = 11.46TRR8 pKa = 11.84ATPWKK13 pKa = 9.08QYY15 pKa = 9.7CFKK18 pKa = 10.94SFPGWYY24 pKa = 9.62KK25 pKa = 10.3VDD27 pKa = 4.05YY28 pKa = 9.96PSSTYY33 pKa = 9.85IDD35 pKa = 3.8EE36 pKa = 4.44EE37 pKa = 4.32VSLRR41 pKa = 11.84NRR43 pKa = 11.84ILLPMPQPQLNTPQWLSFVRR63 pKa = 11.84EE64 pKa = 3.99MRR66 pKa = 11.84HH67 pKa = 6.14LKK69 pKa = 9.57HH70 pKa = 5.69QVPIVEE76 pKa = 4.4TLTRR80 pKa = 11.84QQVILKK86 pKa = 7.68YY87 pKa = 8.92TGAKK91 pKa = 6.64RR92 pKa = 11.84TRR94 pKa = 11.84YY95 pKa = 8.64EE96 pKa = 3.61KK97 pKa = 10.83AAISLMTKK105 pKa = 9.9PLNKK109 pKa = 8.91RR110 pKa = 11.84DD111 pKa = 3.68SYY113 pKa = 11.28IDD115 pKa = 3.63CFLKK119 pKa = 10.43VEE121 pKa = 4.31KK122 pKa = 9.93MPHH125 pKa = 4.82EE126 pKa = 4.1TLKK129 pKa = 10.7IKK131 pKa = 10.45APRR134 pKa = 11.84VIQGRR139 pKa = 11.84SPRR142 pKa = 11.84YY143 pKa = 8.28NLEE146 pKa = 3.58LMRR149 pKa = 11.84YY150 pKa = 8.46LRR152 pKa = 11.84PIEE155 pKa = 3.59HH156 pKa = 7.11WFYY159 pKa = 11.16QKK161 pKa = 11.0PEE163 pKa = 3.83ILKK166 pKa = 9.49TNSLEE171 pKa = 3.91QRR173 pKa = 11.84ALKK176 pKa = 9.91IQQLLDD182 pKa = 3.17KK183 pKa = 10.16HH184 pKa = 6.91DD185 pKa = 4.2NPTVYY190 pKa = 10.52QIDD193 pKa = 3.96CSKK196 pKa = 11.0CDD198 pKa = 3.02AHH200 pKa = 5.75TTHH203 pKa = 6.61HH204 pKa = 5.92VLRR207 pKa = 11.84LEE209 pKa = 3.9HH210 pKa = 6.82NIYY213 pKa = 10.61KK214 pKa = 10.27KK215 pKa = 10.71ASRR218 pKa = 11.84NDD220 pKa = 3.56AKK222 pKa = 10.52CSQMLDD228 pKa = 3.22WQLLNVGFTPNGWTYY243 pKa = 9.46KK244 pKa = 7.51TTGRR248 pKa = 11.84RR249 pKa = 11.84ASGDD253 pKa = 3.53FNTGLGTSIICRR265 pKa = 11.84SLLKK269 pKa = 10.87GCIKK273 pKa = 9.39YY274 pKa = 10.15HH275 pKa = 6.28RR276 pKa = 11.84LRR278 pKa = 11.84ASFVCDD284 pKa = 4.04GDD286 pKa = 3.89DD287 pKa = 4.55CMLITSGKK295 pKa = 7.7LTRR298 pKa = 11.84AQIEE302 pKa = 4.18QVRR305 pKa = 11.84YY306 pKa = 9.78YY307 pKa = 10.75YY308 pKa = 9.65LQCGYY313 pKa = 10.63EE314 pKa = 3.88ITLDD318 pKa = 3.18EE319 pKa = 4.53CEE321 pKa = 4.44NYY323 pKa = 10.36EE324 pKa = 4.65EE325 pKa = 5.94AIHH328 pKa = 6.31CQTALIKK335 pKa = 9.74TPTPIMVRR343 pKa = 11.84KK344 pKa = 8.77PWDD347 pKa = 3.97TISKK351 pKa = 10.41CMVSQKK357 pKa = 10.62YY358 pKa = 7.56FTDD361 pKa = 3.3SKK363 pKa = 9.03TASAFLHH370 pKa = 6.04QMATCEE376 pKa = 4.02LSIHH380 pKa = 6.32RR381 pKa = 11.84GVPIMQSFFRR391 pKa = 11.84MILRR395 pKa = 11.84STKK398 pKa = 9.89PNNRR402 pKa = 11.84NYY404 pKa = 10.61SDD406 pKa = 3.96EE407 pKa = 4.2FQHH410 pKa = 7.2RR411 pKa = 11.84LVLVKK416 pKa = 8.99QLCDD420 pKa = 3.16NRR422 pKa = 11.84QDD424 pKa = 3.03ITTEE428 pKa = 3.56ARR430 pKa = 11.84ISFEE434 pKa = 4.4LSFKK438 pKa = 10.94VDD440 pKa = 2.72IATQYY445 pKa = 9.14YY446 pKa = 8.07WEE448 pKa = 4.34EE449 pKa = 4.12VFDD452 pKa = 3.69NWKK455 pKa = 10.33LDD457 pKa = 3.31TSIFDD462 pKa = 3.63SQHH465 pKa = 4.74
MM1 pKa = 7.67SEE3 pKa = 3.95RR4 pKa = 11.84DD5 pKa = 3.42KK6 pKa = 11.46TRR8 pKa = 11.84ATPWKK13 pKa = 9.08QYY15 pKa = 9.7CFKK18 pKa = 10.94SFPGWYY24 pKa = 9.62KK25 pKa = 10.3VDD27 pKa = 4.05YY28 pKa = 9.96PSSTYY33 pKa = 9.85IDD35 pKa = 3.8EE36 pKa = 4.44EE37 pKa = 4.32VSLRR41 pKa = 11.84NRR43 pKa = 11.84ILLPMPQPQLNTPQWLSFVRR63 pKa = 11.84EE64 pKa = 3.99MRR66 pKa = 11.84HH67 pKa = 6.14LKK69 pKa = 9.57HH70 pKa = 5.69QVPIVEE76 pKa = 4.4TLTRR80 pKa = 11.84QQVILKK86 pKa = 7.68YY87 pKa = 8.92TGAKK91 pKa = 6.64RR92 pKa = 11.84TRR94 pKa = 11.84YY95 pKa = 8.64EE96 pKa = 3.61KK97 pKa = 10.83AAISLMTKK105 pKa = 9.9PLNKK109 pKa = 8.91RR110 pKa = 11.84DD111 pKa = 3.68SYY113 pKa = 11.28IDD115 pKa = 3.63CFLKK119 pKa = 10.43VEE121 pKa = 4.31KK122 pKa = 9.93MPHH125 pKa = 4.82EE126 pKa = 4.1TLKK129 pKa = 10.7IKK131 pKa = 10.45APRR134 pKa = 11.84VIQGRR139 pKa = 11.84SPRR142 pKa = 11.84YY143 pKa = 8.28NLEE146 pKa = 3.58LMRR149 pKa = 11.84YY150 pKa = 8.46LRR152 pKa = 11.84PIEE155 pKa = 3.59HH156 pKa = 7.11WFYY159 pKa = 11.16QKK161 pKa = 11.0PEE163 pKa = 3.83ILKK166 pKa = 9.49TNSLEE171 pKa = 3.91QRR173 pKa = 11.84ALKK176 pKa = 9.91IQQLLDD182 pKa = 3.17KK183 pKa = 10.16HH184 pKa = 6.91DD185 pKa = 4.2NPTVYY190 pKa = 10.52QIDD193 pKa = 3.96CSKK196 pKa = 11.0CDD198 pKa = 3.02AHH200 pKa = 5.75TTHH203 pKa = 6.61HH204 pKa = 5.92VLRR207 pKa = 11.84LEE209 pKa = 3.9HH210 pKa = 6.82NIYY213 pKa = 10.61KK214 pKa = 10.27KK215 pKa = 10.71ASRR218 pKa = 11.84NDD220 pKa = 3.56AKK222 pKa = 10.52CSQMLDD228 pKa = 3.22WQLLNVGFTPNGWTYY243 pKa = 9.46KK244 pKa = 7.51TTGRR248 pKa = 11.84RR249 pKa = 11.84ASGDD253 pKa = 3.53FNTGLGTSIICRR265 pKa = 11.84SLLKK269 pKa = 10.87GCIKK273 pKa = 9.39YY274 pKa = 10.15HH275 pKa = 6.28RR276 pKa = 11.84LRR278 pKa = 11.84ASFVCDD284 pKa = 4.04GDD286 pKa = 3.89DD287 pKa = 4.55CMLITSGKK295 pKa = 7.7LTRR298 pKa = 11.84AQIEE302 pKa = 4.18QVRR305 pKa = 11.84YY306 pKa = 9.78YY307 pKa = 10.75YY308 pKa = 9.65LQCGYY313 pKa = 10.63EE314 pKa = 3.88ITLDD318 pKa = 3.18EE319 pKa = 4.53CEE321 pKa = 4.44NYY323 pKa = 10.36EE324 pKa = 4.65EE325 pKa = 5.94AIHH328 pKa = 6.31CQTALIKK335 pKa = 9.74TPTPIMVRR343 pKa = 11.84KK344 pKa = 8.77PWDD347 pKa = 3.97TISKK351 pKa = 10.41CMVSQKK357 pKa = 10.62YY358 pKa = 7.56FTDD361 pKa = 3.3SKK363 pKa = 9.03TASAFLHH370 pKa = 6.04QMATCEE376 pKa = 4.02LSIHH380 pKa = 6.32RR381 pKa = 11.84GVPIMQSFFRR391 pKa = 11.84MILRR395 pKa = 11.84STKK398 pKa = 9.89PNNRR402 pKa = 11.84NYY404 pKa = 10.61SDD406 pKa = 3.96EE407 pKa = 4.2FQHH410 pKa = 7.2RR411 pKa = 11.84LVLVKK416 pKa = 8.99QLCDD420 pKa = 3.16NRR422 pKa = 11.84QDD424 pKa = 3.03ITTEE428 pKa = 3.56ARR430 pKa = 11.84ISFEE434 pKa = 4.4LSFKK438 pKa = 10.94VDD440 pKa = 2.72IATQYY445 pKa = 9.14YY446 pKa = 8.07WEE448 pKa = 4.34EE449 pKa = 4.12VFDD452 pKa = 3.69NWKK455 pKa = 10.33LDD457 pKa = 3.31TSIFDD462 pKa = 3.63SQHH465 pKa = 4.74
Molecular weight: 54.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
720 |
255 |
465 |
360.0 |
41.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.278 ± 1.054 | 2.5 ± 0.642 |
5.0 ± 0.333 | 5.833 ± 0.404 |
3.472 ± 0.162 | 3.611 ± 0.531 |
3.194 ± 0.028 | 5.694 ± 0.67 |
6.528 ± 0.884 | 9.167 ± 0.119 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.016 | 4.028 ± 0.329 |
5.417 ± 0.796 | 5.972 ± 0.147 |
6.528 ± 0.503 | 6.944 ± 0.436 |
7.083 ± 0.392 | 4.722 ± 0.372 |
1.667 ± 0.238 | 4.583 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
