
Durio zibethinus (Durian)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Helicteroideae; Durio
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
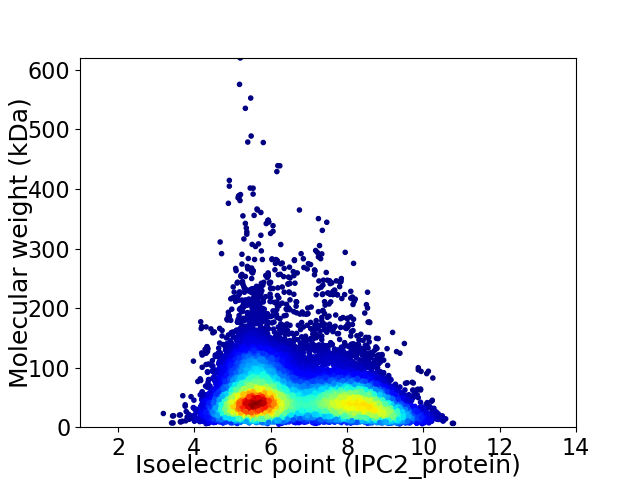
Virtual 2D-PAGE plot for 53026 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P5XK65|A0A6P5XK65_DURZI benzyl alcohol O-benzoyltransferase-like OS=Durio zibethinus OX=66656 GN=LOC111284102 PE=3 SV=1
MM1 pKa = 7.88PKK3 pKa = 10.39FNFPTILILAILLLACEE20 pKa = 4.48LAIISPVLAYY30 pKa = 10.88NDD32 pKa = 3.39VDD34 pKa = 4.01IGDD37 pKa = 4.27ISPSAVTDD45 pKa = 3.57DD46 pKa = 5.56DD47 pKa = 4.92MITSSSLDD55 pKa = 3.31AGQLEE60 pKa = 4.51DD61 pKa = 4.52LYY63 pKa = 10.89QRR65 pKa = 11.84CVVMLGDD72 pKa = 3.6EE73 pKa = 4.62CGDD76 pKa = 3.97EE77 pKa = 4.08IFDD80 pKa = 5.49LIFRR84 pKa = 11.84NDD86 pKa = 3.32SLIVTDD92 pKa = 4.7KK93 pKa = 11.35DD94 pKa = 2.86IGLVSKK100 pKa = 8.89YY101 pKa = 10.88CCDD104 pKa = 3.21KK105 pKa = 10.64LLNMGRR111 pKa = 11.84NCHH114 pKa = 6.01DD115 pKa = 3.19QLVNVIAQTTHH126 pKa = 6.53FSTNSSATVPRR137 pKa = 11.84SSQVWDD143 pKa = 3.69LCSLSSSS150 pKa = 3.53
MM1 pKa = 7.88PKK3 pKa = 10.39FNFPTILILAILLLACEE20 pKa = 4.48LAIISPVLAYY30 pKa = 10.88NDD32 pKa = 3.39VDD34 pKa = 4.01IGDD37 pKa = 4.27ISPSAVTDD45 pKa = 3.57DD46 pKa = 5.56DD47 pKa = 4.92MITSSSLDD55 pKa = 3.31AGQLEE60 pKa = 4.51DD61 pKa = 4.52LYY63 pKa = 10.89QRR65 pKa = 11.84CVVMLGDD72 pKa = 3.6EE73 pKa = 4.62CGDD76 pKa = 3.97EE77 pKa = 4.08IFDD80 pKa = 5.49LIFRR84 pKa = 11.84NDD86 pKa = 3.32SLIVTDD92 pKa = 4.7KK93 pKa = 11.35DD94 pKa = 2.86IGLVSKK100 pKa = 8.89YY101 pKa = 10.88CCDD104 pKa = 3.21KK105 pKa = 10.64LLNMGRR111 pKa = 11.84NCHH114 pKa = 6.01DD115 pKa = 3.19QLVNVIAQTTHH126 pKa = 6.53FSTNSSATVPRR137 pKa = 11.84SSQVWDD143 pKa = 3.69LCSLSSSS150 pKa = 3.53
Molecular weight: 16.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P5ZWJ5|A0A6P5ZWJ5_DURZI Peptidyl-prolyl cis-trans isomerase OS=Durio zibethinus OX=66656 GN=LOC111304638 PE=3 SV=1
MM1 pKa = 6.7YY2 pKa = 10.1HH3 pKa = 6.64SLRR6 pKa = 11.84SQSLSPRR13 pKa = 11.84SCFPARR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84PSYY24 pKa = 10.1SRR26 pKa = 11.84QRR28 pKa = 11.84SRR30 pKa = 11.84SHH32 pKa = 5.84SLSRR36 pKa = 11.84SPSPVRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84LRR46 pKa = 11.84SPYY49 pKa = 9.74QRR51 pKa = 11.84WSPTLVRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 9.42LSPICRR67 pKa = 11.84HH68 pKa = 6.2RR69 pKa = 11.84SPLSIRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84SSSLVRR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 6.21RR87 pKa = 11.84SPSTVFRR94 pKa = 11.84RR95 pKa = 11.84SSSPVRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84SASSVCRR110 pKa = 11.84RR111 pKa = 11.84SPLPMCRR118 pKa = 11.84RR119 pKa = 11.84SPSPLRR125 pKa = 11.84HH126 pKa = 6.35RR127 pKa = 11.84SPHH130 pKa = 4.11MQRR133 pKa = 11.84RR134 pKa = 11.84SPSSVRR140 pKa = 11.84QRR142 pKa = 11.84HH143 pKa = 5.51HH144 pKa = 6.95RR145 pKa = 11.84LSSTPRR151 pKa = 11.84HH152 pKa = 5.86RR153 pKa = 11.84SSSLVRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84SPGSNRR167 pKa = 11.84RR168 pKa = 11.84SITPSHH174 pKa = 6.23GKK176 pKa = 9.56SLSLYY181 pKa = 8.9QSCSLSPVQHH191 pKa = 6.7RR192 pKa = 11.84SSSPLKK198 pKa = 10.41RR199 pKa = 11.84SPKK202 pKa = 10.17DD203 pKa = 3.09
MM1 pKa = 6.7YY2 pKa = 10.1HH3 pKa = 6.64SLRR6 pKa = 11.84SQSLSPRR13 pKa = 11.84SCFPARR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84PSYY24 pKa = 10.1SRR26 pKa = 11.84QRR28 pKa = 11.84SRR30 pKa = 11.84SHH32 pKa = 5.84SLSRR36 pKa = 11.84SPSPVRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84LRR46 pKa = 11.84SPYY49 pKa = 9.74QRR51 pKa = 11.84WSPTLVRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 9.42LSPICRR67 pKa = 11.84HH68 pKa = 6.2RR69 pKa = 11.84SPLSIRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84SSSLVRR84 pKa = 11.84RR85 pKa = 11.84HH86 pKa = 6.21RR87 pKa = 11.84SPSTVFRR94 pKa = 11.84RR95 pKa = 11.84SSSPVRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84SASSVCRR110 pKa = 11.84RR111 pKa = 11.84SPLPMCRR118 pKa = 11.84RR119 pKa = 11.84SPSPLRR125 pKa = 11.84HH126 pKa = 6.35RR127 pKa = 11.84SPHH130 pKa = 4.11MQRR133 pKa = 11.84RR134 pKa = 11.84SPSSVRR140 pKa = 11.84QRR142 pKa = 11.84HH143 pKa = 5.51HH144 pKa = 6.95RR145 pKa = 11.84LSSTPRR151 pKa = 11.84HH152 pKa = 5.86RR153 pKa = 11.84SSSLVRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84SPGSNRR167 pKa = 11.84RR168 pKa = 11.84SITPSHH174 pKa = 6.23GKK176 pKa = 9.56SLSLYY181 pKa = 8.9QSCSLSPVQHH191 pKa = 6.7RR192 pKa = 11.84SSSPLKK198 pKa = 10.41RR199 pKa = 11.84SPKK202 pKa = 10.17DD203 pKa = 3.09
Molecular weight: 23.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
26388952 |
31 |
5460 |
497.7 |
55.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.651 ± 0.008 | 1.882 ± 0.005 |
5.28 ± 0.007 | 6.555 ± 0.012 |
4.217 ± 0.006 | 6.406 ± 0.01 |
2.371 ± 0.004 | 5.349 ± 0.008 |
6.135 ± 0.01 | 9.934 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.382 ± 0.003 | 4.574 ± 0.007 |
4.835 ± 0.009 | 3.881 ± 0.008 |
5.209 ± 0.008 | 9.231 ± 0.012 |
4.736 ± 0.006 | 6.386 ± 0.007 |
1.245 ± 0.003 | 2.738 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
