
Maize yellow dwarf virus-RMV2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; Maize yellow mosaic virus
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
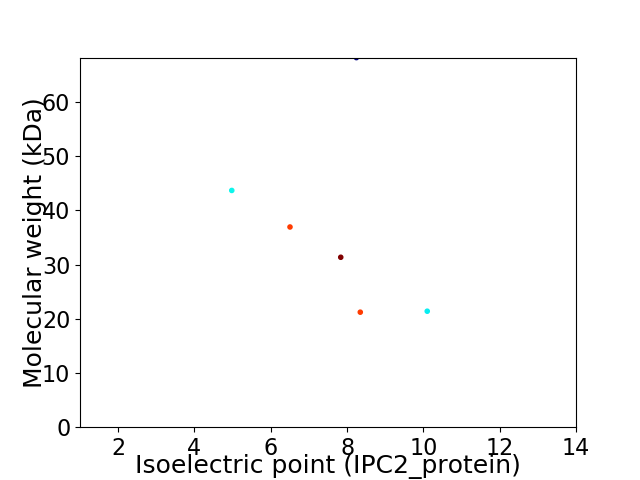
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165G9A9|A0A165G9A9_9LUTE p5 OS=Maize yellow dwarf virus-RMV2 OX=1833824 PE=4 SV=1
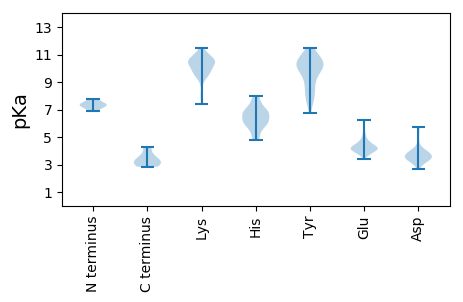
MM1 pKa = 7.33YY2 pKa = 9.85YY3 pKa = 9.27WHH5 pKa = 7.62DD6 pKa = 3.83EE7 pKa = 4.19SWSKK11 pKa = 9.2EE12 pKa = 4.25TLSAGYY18 pKa = 8.96VQNDD22 pKa = 3.46SSRR25 pKa = 11.84ATPYY29 pKa = 10.56FLIPTVVGNYY39 pKa = 8.95KK40 pKa = 10.44VYY42 pKa = 10.37IEE44 pKa = 4.78CEE46 pKa = 3.72GFQAVKK52 pKa = 10.8AKK54 pKa = 10.66GGEE57 pKa = 4.32NNGKK61 pKa = 8.12MAGFITYY68 pKa = 10.0DD69 pKa = 3.48PQQAGWQAYY78 pKa = 7.54SWAGCTLSNYY88 pKa = 10.07RR89 pKa = 11.84ITDD92 pKa = 3.52GCVQAHH98 pKa = 7.54PDD100 pKa = 3.81LKK102 pKa = 11.51VNGCSFTKK110 pKa = 9.81GQCVEE115 pKa = 4.11RR116 pKa = 11.84DD117 pKa = 3.61VVISFDD123 pKa = 3.85LSVASEE129 pKa = 5.05GYY131 pKa = 8.45WALQAPPIEE140 pKa = 4.59KK141 pKa = 9.74TDD143 pKa = 3.37DD144 pKa = 3.33HH145 pKa = 7.69NFIVSYY151 pKa = 11.25GNYY154 pKa = 7.86TEE156 pKa = 6.16KK157 pKa = 10.44ILEE160 pKa = 4.04WGMVSISIDD169 pKa = 3.9EE170 pKa = 4.37INSSNDD176 pKa = 2.82AQRR179 pKa = 11.84VPNRR183 pKa = 11.84NKK185 pKa = 9.79DD186 pKa = 3.23TSEE189 pKa = 4.03PNRR192 pKa = 11.84LTDD195 pKa = 3.89FTNSASVGVTVTDD208 pKa = 3.9KK209 pKa = 11.36NLDD212 pKa = 3.85SEE214 pKa = 4.88PEE216 pKa = 4.06GQLVVQTPSLQTPANPVAEE235 pKa = 4.44PASKK239 pKa = 10.8VSAKK243 pKa = 9.83VVAWLNNPDD252 pKa = 4.17PEE254 pKa = 5.01SDD256 pKa = 3.73ADD258 pKa = 4.01APPPPTPWDD267 pKa = 3.48SSQVRR272 pKa = 11.84LRR274 pKa = 11.84SRR276 pKa = 11.84EE277 pKa = 3.87EE278 pKa = 3.88ANLEE282 pKa = 3.93PSPQNASPRR291 pKa = 11.84EE292 pKa = 4.15YY293 pKa = 10.24MNKK296 pKa = 9.34TYY298 pKa = 10.69EE299 pKa = 4.69DD300 pKa = 4.27DD301 pKa = 3.9VASQATKK308 pKa = 10.27TPTLSGGSLRR318 pKa = 11.84PRR320 pKa = 11.84LKK322 pKa = 9.45QTSDD326 pKa = 3.09SAKK329 pKa = 10.65DD330 pKa = 3.37NSSQVGSQTGTLTGGSLRR348 pKa = 11.84PSLQGGRR355 pKa = 11.84LASQQFRR362 pKa = 11.84MTTEE366 pKa = 3.44QAKK369 pKa = 10.4QYY371 pKa = 11.12DD372 pKa = 4.33LIKK375 pKa = 10.51RR376 pKa = 11.84SMGRR380 pKa = 11.84TRR382 pKa = 11.84ANEE385 pKa = 3.88YY386 pKa = 10.78YY387 pKa = 10.32EE388 pKa = 4.41EE389 pKa = 4.14CRR391 pKa = 11.84KK392 pKa = 10.01AAEE395 pKa = 4.43GSS397 pKa = 3.55
MM1 pKa = 7.33YY2 pKa = 9.85YY3 pKa = 9.27WHH5 pKa = 7.62DD6 pKa = 3.83EE7 pKa = 4.19SWSKK11 pKa = 9.2EE12 pKa = 4.25TLSAGYY18 pKa = 8.96VQNDD22 pKa = 3.46SSRR25 pKa = 11.84ATPYY29 pKa = 10.56FLIPTVVGNYY39 pKa = 8.95KK40 pKa = 10.44VYY42 pKa = 10.37IEE44 pKa = 4.78CEE46 pKa = 3.72GFQAVKK52 pKa = 10.8AKK54 pKa = 10.66GGEE57 pKa = 4.32NNGKK61 pKa = 8.12MAGFITYY68 pKa = 10.0DD69 pKa = 3.48PQQAGWQAYY78 pKa = 7.54SWAGCTLSNYY88 pKa = 10.07RR89 pKa = 11.84ITDD92 pKa = 3.52GCVQAHH98 pKa = 7.54PDD100 pKa = 3.81LKK102 pKa = 11.51VNGCSFTKK110 pKa = 9.81GQCVEE115 pKa = 4.11RR116 pKa = 11.84DD117 pKa = 3.61VVISFDD123 pKa = 3.85LSVASEE129 pKa = 5.05GYY131 pKa = 8.45WALQAPPIEE140 pKa = 4.59KK141 pKa = 9.74TDD143 pKa = 3.37DD144 pKa = 3.33HH145 pKa = 7.69NFIVSYY151 pKa = 11.25GNYY154 pKa = 7.86TEE156 pKa = 6.16KK157 pKa = 10.44ILEE160 pKa = 4.04WGMVSISIDD169 pKa = 3.9EE170 pKa = 4.37INSSNDD176 pKa = 2.82AQRR179 pKa = 11.84VPNRR183 pKa = 11.84NKK185 pKa = 9.79DD186 pKa = 3.23TSEE189 pKa = 4.03PNRR192 pKa = 11.84LTDD195 pKa = 3.89FTNSASVGVTVTDD208 pKa = 3.9KK209 pKa = 11.36NLDD212 pKa = 3.85SEE214 pKa = 4.88PEE216 pKa = 4.06GQLVVQTPSLQTPANPVAEE235 pKa = 4.44PASKK239 pKa = 10.8VSAKK243 pKa = 9.83VVAWLNNPDD252 pKa = 4.17PEE254 pKa = 5.01SDD256 pKa = 3.73ADD258 pKa = 4.01APPPPTPWDD267 pKa = 3.48SSQVRR272 pKa = 11.84LRR274 pKa = 11.84SRR276 pKa = 11.84EE277 pKa = 3.87EE278 pKa = 3.88ANLEE282 pKa = 3.93PSPQNASPRR291 pKa = 11.84EE292 pKa = 4.15YY293 pKa = 10.24MNKK296 pKa = 9.34TYY298 pKa = 10.69EE299 pKa = 4.69DD300 pKa = 4.27DD301 pKa = 3.9VASQATKK308 pKa = 10.27TPTLSGGSLRR318 pKa = 11.84PRR320 pKa = 11.84LKK322 pKa = 9.45QTSDD326 pKa = 3.09SAKK329 pKa = 10.65DD330 pKa = 3.37NSSQVGSQTGTLTGGSLRR348 pKa = 11.84PSLQGGRR355 pKa = 11.84LASQQFRR362 pKa = 11.84MTTEE366 pKa = 3.44QAKK369 pKa = 10.4QYY371 pKa = 11.12DD372 pKa = 4.33LIKK375 pKa = 10.51RR376 pKa = 11.84SMGRR380 pKa = 11.84TRR382 pKa = 11.84ANEE385 pKa = 3.88YY386 pKa = 10.78YY387 pKa = 10.32EE388 pKa = 4.41EE389 pKa = 4.14CRR391 pKa = 11.84KK392 pKa = 10.01AAEE395 pKa = 4.43GSS397 pKa = 3.55
Molecular weight: 43.68 kDa
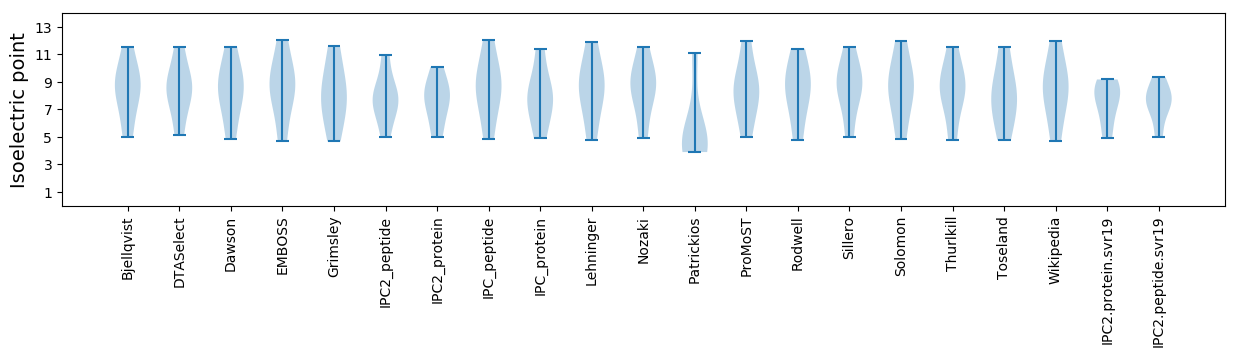
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165G999|A0A165G999_9LUTE Movement protein OS=Maize yellow dwarf virus-RMV2 OX=1833824 PE=3 SV=1
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84ARR18 pKa = 11.84NNNRR22 pKa = 11.84AQPVVVVAANPRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PSGNTAGRR51 pKa = 11.84PGVRR55 pKa = 11.84RR56 pKa = 11.84GPRR59 pKa = 11.84EE60 pKa = 3.62TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 7.79VTFGPSLSEE85 pKa = 3.84CAAFSGGILKK95 pKa = 10.27AYY97 pKa = 10.08HH98 pKa = 6.52EE99 pKa = 4.39YY100 pKa = 10.12KK101 pKa = 10.44IKK103 pKa = 10.53IILEE107 pKa = 4.7FISEE111 pKa = 4.21AASTAEE117 pKa = 3.73GSIAYY122 pKa = 9.5EE123 pKa = 3.8LDD125 pKa = 3.19PHH127 pKa = 6.74NKK129 pKa = 9.68LSSLASTINKK139 pKa = 9.5FSIVKK144 pKa = 9.57GGRR147 pKa = 11.84RR148 pKa = 11.84VFASNQIGGGVWRR161 pKa = 11.84DD162 pKa = 3.39SSEE165 pKa = 4.18DD166 pKa = 3.19QCTIHH171 pKa = 6.52YY172 pKa = 9.09KK173 pKa = 11.03GNGKK177 pKa = 10.18SSVAGSFRR185 pKa = 11.84ITIEE189 pKa = 4.01VNVQNPRR196 pKa = 3.21
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84ARR18 pKa = 11.84NNNRR22 pKa = 11.84AQPVVVVAANPRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PSGNTAGRR51 pKa = 11.84PGVRR55 pKa = 11.84RR56 pKa = 11.84GPRR59 pKa = 11.84EE60 pKa = 3.62TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 7.79VTFGPSLSEE85 pKa = 3.84CAAFSGGILKK95 pKa = 10.27AYY97 pKa = 10.08HH98 pKa = 6.52EE99 pKa = 4.39YY100 pKa = 10.12KK101 pKa = 10.44IKK103 pKa = 10.53IILEE107 pKa = 4.7FISEE111 pKa = 4.21AASTAEE117 pKa = 3.73GSIAYY122 pKa = 9.5EE123 pKa = 3.8LDD125 pKa = 3.19PHH127 pKa = 6.74NKK129 pKa = 9.68LSSLASTINKK139 pKa = 9.5FSIVKK144 pKa = 9.57GGRR147 pKa = 11.84RR148 pKa = 11.84VFASNQIGGGVWRR161 pKa = 11.84DD162 pKa = 3.39SSEE165 pKa = 4.18DD166 pKa = 3.19QCTIHH171 pKa = 6.52YY172 pKa = 9.09KK173 pKa = 11.03GNGKK177 pKa = 10.18SSVAGSFRR185 pKa = 11.84ITIEE189 pKa = 4.01VNVQNPRR196 pKa = 3.21
Molecular weight: 21.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2004 |
191 |
622 |
334.0 |
37.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.886 ± 0.41 | 1.747 ± 0.301 |
4.691 ± 0.711 | 5.439 ± 0.408 |
3.792 ± 0.731 | 6.986 ± 0.818 |
1.996 ± 0.412 | 3.992 ± 0.52 |
4.591 ± 0.573 | 9.481 ± 1.4 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.747 ± 0.328 | 4.341 ± 0.771 |
5.639 ± 0.685 | 4.341 ± 0.645 |
7.036 ± 1.441 | 10.08 ± 0.822 |
6.088 ± 0.59 | 6.487 ± 0.607 |
1.547 ± 0.235 | 3.094 ± 0.389 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
