
Bacillus phage Palmer
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Pagevirus; Bacillus virus Palmer
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
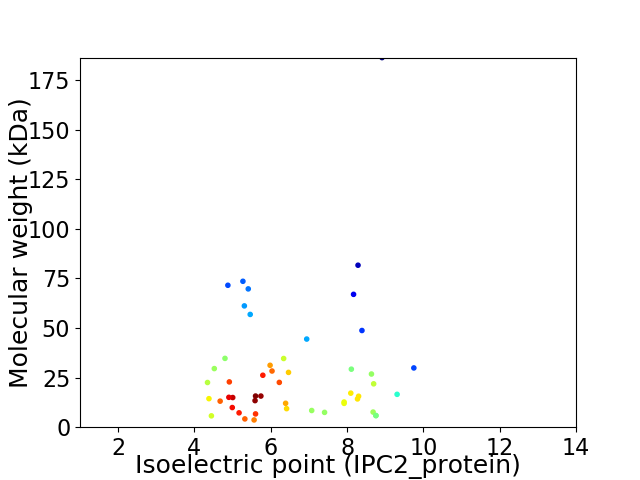
Virtual 2D-PAGE plot for 50 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5AFI4|A0A0C5AFI4_9CAUD Uncharacterized protein OS=Bacillus phage Palmer OX=1597966 GN=CPT_Palmer45 PE=4 SV=1
MM1 pKa = 7.32NLGEE5 pKa = 4.62AKK7 pKa = 10.34AHH9 pKa = 6.22ALALMAEE16 pKa = 4.16YY17 pKa = 10.45SIDD20 pKa = 3.45GLEE23 pKa = 4.55ISDD26 pKa = 5.36AEE28 pKa = 4.07NADD31 pKa = 3.5YY32 pKa = 10.84LKK34 pKa = 10.85RR35 pKa = 11.84MPLFAHH41 pKa = 7.09EE42 pKa = 4.19AQMDD46 pKa = 3.72ISARR50 pKa = 11.84SGVTAVMSFEE60 pKa = 4.81LVKK63 pKa = 10.66QDD65 pKa = 3.55DD66 pKa = 3.62VKK68 pKa = 11.45YY69 pKa = 11.14NYY71 pKa = 9.35IALPADD77 pKa = 3.74YY78 pKa = 10.93RR79 pKa = 11.84EE80 pKa = 4.09MKK82 pKa = 9.89YY83 pKa = 10.14VFWNEE88 pKa = 3.37EE89 pKa = 4.35LFTGNYY95 pKa = 10.01SILNNQIRR103 pKa = 11.84FNAGFDD109 pKa = 3.67GTIDD113 pKa = 3.92LYY115 pKa = 9.91YY116 pKa = 10.12WKK118 pKa = 10.42YY119 pKa = 8.95PQQLTTQTEE128 pKa = 3.81DD129 pKa = 3.35DD130 pKa = 4.27YY131 pKa = 11.74EE132 pKa = 4.44FEE134 pKa = 4.15IDD136 pKa = 4.39RR137 pKa = 11.84EE138 pKa = 4.24YY139 pKa = 11.32HH140 pKa = 6.37HH141 pKa = 7.26LIPYY145 pKa = 9.22YY146 pKa = 10.75LGGKK150 pKa = 9.44CMQDD154 pKa = 3.11EE155 pKa = 4.36NLDD158 pKa = 3.33IAEE161 pKa = 4.77RR162 pKa = 11.84LLSDD166 pKa = 3.7YY167 pKa = 11.02YY168 pKa = 11.67NRR170 pKa = 11.84LSDD173 pKa = 3.48VKK175 pKa = 10.15EE176 pKa = 3.77AHH178 pKa = 6.97EE179 pKa = 5.11DD180 pKa = 3.5GQEE183 pKa = 4.06RR184 pKa = 11.84IDD186 pKa = 4.04NIFTIFF192 pKa = 3.35
MM1 pKa = 7.32NLGEE5 pKa = 4.62AKK7 pKa = 10.34AHH9 pKa = 6.22ALALMAEE16 pKa = 4.16YY17 pKa = 10.45SIDD20 pKa = 3.45GLEE23 pKa = 4.55ISDD26 pKa = 5.36AEE28 pKa = 4.07NADD31 pKa = 3.5YY32 pKa = 10.84LKK34 pKa = 10.85RR35 pKa = 11.84MPLFAHH41 pKa = 7.09EE42 pKa = 4.19AQMDD46 pKa = 3.72ISARR50 pKa = 11.84SGVTAVMSFEE60 pKa = 4.81LVKK63 pKa = 10.66QDD65 pKa = 3.55DD66 pKa = 3.62VKK68 pKa = 11.45YY69 pKa = 11.14NYY71 pKa = 9.35IALPADD77 pKa = 3.74YY78 pKa = 10.93RR79 pKa = 11.84EE80 pKa = 4.09MKK82 pKa = 9.89YY83 pKa = 10.14VFWNEE88 pKa = 3.37EE89 pKa = 4.35LFTGNYY95 pKa = 10.01SILNNQIRR103 pKa = 11.84FNAGFDD109 pKa = 3.67GTIDD113 pKa = 3.92LYY115 pKa = 9.91YY116 pKa = 10.12WKK118 pKa = 10.42YY119 pKa = 8.95PQQLTTQTEE128 pKa = 3.81DD129 pKa = 3.35DD130 pKa = 4.27YY131 pKa = 11.74EE132 pKa = 4.44FEE134 pKa = 4.15IDD136 pKa = 4.39RR137 pKa = 11.84EE138 pKa = 4.24YY139 pKa = 11.32HH140 pKa = 6.37HH141 pKa = 7.26LIPYY145 pKa = 9.22YY146 pKa = 10.75LGGKK150 pKa = 9.44CMQDD154 pKa = 3.11EE155 pKa = 4.36NLDD158 pKa = 3.33IAEE161 pKa = 4.77RR162 pKa = 11.84LLSDD166 pKa = 3.7YY167 pKa = 11.02YY168 pKa = 11.67NRR170 pKa = 11.84LSDD173 pKa = 3.48VKK175 pKa = 10.15EE176 pKa = 3.77AHH178 pKa = 6.97EE179 pKa = 5.11DD180 pKa = 3.5GQEE183 pKa = 4.06RR184 pKa = 11.84IDD186 pKa = 4.04NIFTIFF192 pKa = 3.35
Molecular weight: 22.53 kDa
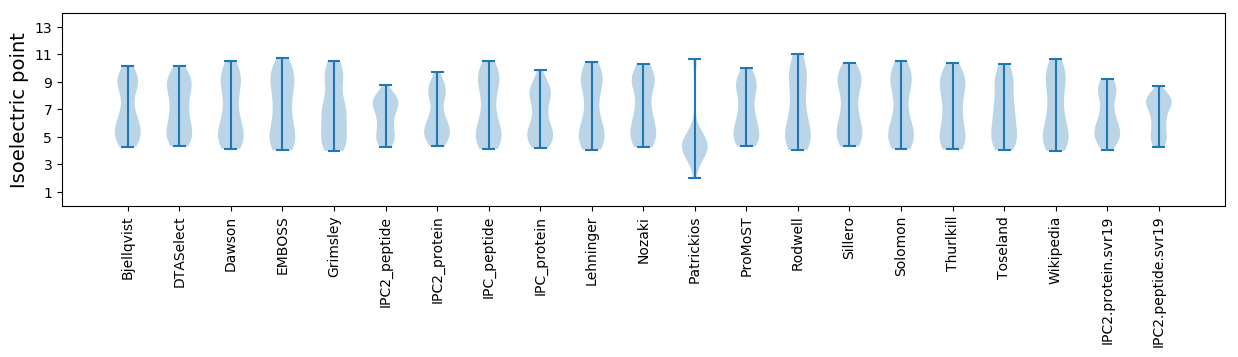
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5ACE6|A0A0C5ACE6_9CAUD Uncharacterized protein OS=Bacillus phage Palmer OX=1597966 GN=CPT_Palmer24 PE=4 SV=1
MM1 pKa = 7.31FGGGRR6 pKa = 11.84SEE8 pKa = 4.12KK9 pKa = 10.59DD10 pKa = 2.7IAAGRR15 pKa = 11.84FDD17 pKa = 3.52CSSYY21 pKa = 10.84VRR23 pKa = 11.84WAYY26 pKa = 10.5SQLGINLGPLTSTSTEE42 pKa = 3.87TLKK45 pKa = 11.4NKK47 pKa = 7.98GTAVKK52 pKa = 10.43GGLANAQPGDD62 pKa = 4.19LIFFDD67 pKa = 4.15TYY69 pKa = 10.87KK70 pKa = 11.0KK71 pKa = 10.0NGHH74 pKa = 4.46VVIYY78 pKa = 10.36LGNNQFIGAQGKK90 pKa = 7.34TGVGIVDD97 pKa = 4.61LNSSYY102 pKa = 11.45GSYY105 pKa = 10.07FKK107 pKa = 10.87KK108 pKa = 10.34RR109 pKa = 11.84FSGNIRR115 pKa = 11.84RR116 pKa = 11.84IAGGGNVGAALGGGGAAMSGAGTGTAYY143 pKa = 10.32RR144 pKa = 11.84QAPSNLMGPINNAAKK159 pKa = 10.29QYY161 pKa = 10.27GVNPNLIAAIIKK173 pKa = 10.22KK174 pKa = 10.2EE175 pKa = 4.13STFKK179 pKa = 11.0SGLTSSAGAKK189 pKa = 10.33GYY191 pKa = 7.86MQLMPATARR200 pKa = 11.84AMGVKK205 pKa = 10.22NPWDD209 pKa = 3.58TQQNINGGTKK219 pKa = 10.51YY220 pKa = 9.98IAQQLKK226 pKa = 8.97TYY228 pKa = 10.29KK229 pKa = 10.78NNIPLALAAYY239 pKa = 9.31NWGPGNLNKK248 pKa = 10.5AIRR251 pKa = 11.84KK252 pKa = 9.59AGGSKK257 pKa = 10.02DD258 pKa = 2.92WNQIRR263 pKa = 11.84RR264 pKa = 11.84FAPKK268 pKa = 8.44EE269 pKa = 3.76TRR271 pKa = 11.84DD272 pKa = 3.79YY273 pKa = 11.28VDD275 pKa = 5.67KK276 pKa = 11.37IMGWRR281 pKa = 3.39
MM1 pKa = 7.31FGGGRR6 pKa = 11.84SEE8 pKa = 4.12KK9 pKa = 10.59DD10 pKa = 2.7IAAGRR15 pKa = 11.84FDD17 pKa = 3.52CSSYY21 pKa = 10.84VRR23 pKa = 11.84WAYY26 pKa = 10.5SQLGINLGPLTSTSTEE42 pKa = 3.87TLKK45 pKa = 11.4NKK47 pKa = 7.98GTAVKK52 pKa = 10.43GGLANAQPGDD62 pKa = 4.19LIFFDD67 pKa = 4.15TYY69 pKa = 10.87KK70 pKa = 11.0KK71 pKa = 10.0NGHH74 pKa = 4.46VVIYY78 pKa = 10.36LGNNQFIGAQGKK90 pKa = 7.34TGVGIVDD97 pKa = 4.61LNSSYY102 pKa = 11.45GSYY105 pKa = 10.07FKK107 pKa = 10.87KK108 pKa = 10.34RR109 pKa = 11.84FSGNIRR115 pKa = 11.84RR116 pKa = 11.84IAGGGNVGAALGGGGAAMSGAGTGTAYY143 pKa = 10.32RR144 pKa = 11.84QAPSNLMGPINNAAKK159 pKa = 10.29QYY161 pKa = 10.27GVNPNLIAAIIKK173 pKa = 10.22KK174 pKa = 10.2EE175 pKa = 4.13STFKK179 pKa = 11.0SGLTSSAGAKK189 pKa = 10.33GYY191 pKa = 7.86MQLMPATARR200 pKa = 11.84AMGVKK205 pKa = 10.22NPWDD209 pKa = 3.58TQQNINGGTKK219 pKa = 10.51YY220 pKa = 9.98IAQQLKK226 pKa = 8.97TYY228 pKa = 10.29KK229 pKa = 10.78NNIPLALAAYY239 pKa = 9.31NWGPGNLNKK248 pKa = 10.5AIRR251 pKa = 11.84KK252 pKa = 9.59AGGSKK257 pKa = 10.02DD258 pKa = 2.92WNQIRR263 pKa = 11.84RR264 pKa = 11.84FAPKK268 pKa = 8.44EE269 pKa = 3.76TRR271 pKa = 11.84DD272 pKa = 3.79YY273 pKa = 11.28VDD275 pKa = 5.67KK276 pKa = 11.37IMGWRR281 pKa = 3.39
Molecular weight: 29.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12742 |
30 |
1678 |
254.8 |
28.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.06 ± 0.367 | 0.698 ± 0.135 |
5.556 ± 0.209 | 7.157 ± 0.521 |
4.136 ± 0.172 | 6.906 ± 0.339 |
1.601 ± 0.154 | 6.114 ± 0.395 |
8.28 ± 0.491 | 7.173 ± 0.341 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.833 ± 0.193 | 6.169 ± 0.245 |
3.704 ± 0.36 | 4.881 ± 0.582 |
4.128 ± 0.22 | 5.572 ± 0.23 |
5.972 ± 0.422 | 5.957 ± 0.238 |
1.24 ± 0.119 | 3.861 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
