
Cattle blood-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus cowchi1
Average proteome isoelectric point is 7.84
Get precalculated fractions of proteins
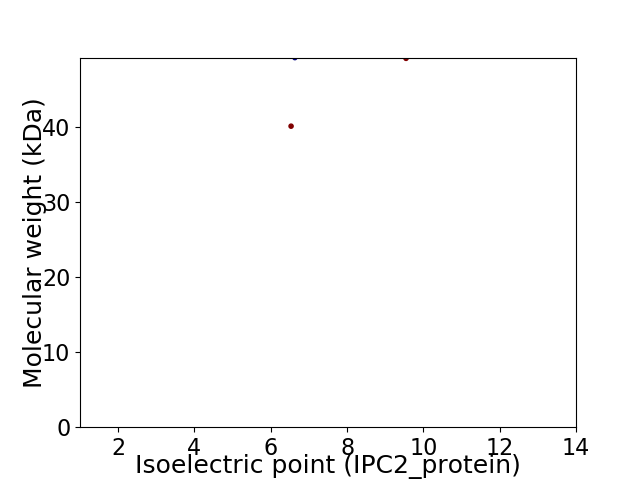
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L0HH91|A0A2L0HH91_9VIRU Replication-associated protein OS=Cattle blood-associated gemycircularvirus OX=2077296 PE=3 SV=1
MM1 pKa = 7.37SQFQFNHH8 pKa = 5.38IQLGLTYY15 pKa = 9.3PQARR19 pKa = 11.84GLTKK23 pKa = 10.26EE24 pKa = 3.91LLRR27 pKa = 11.84IYY29 pKa = 10.69LEE31 pKa = 3.35NWYY34 pKa = 10.08FNNGTCEE41 pKa = 3.96VRR43 pKa = 11.84VTKK46 pKa = 10.46YY47 pKa = 10.66LIAEE51 pKa = 4.34EE52 pKa = 4.04KK53 pKa = 10.83HH54 pKa = 6.26EE55 pKa = 4.74DD56 pKa = 4.0GEE58 pKa = 4.63PHH60 pKa = 5.36FHH62 pKa = 6.74VYY64 pKa = 10.6LRR66 pKa = 11.84FDD68 pKa = 3.19NKK70 pKa = 9.99LRR72 pKa = 11.84ARR74 pKa = 11.84EE75 pKa = 3.89TRR77 pKa = 11.84LFDD80 pKa = 3.76IFFEE84 pKa = 4.69STGKK88 pKa = 10.53LYY90 pKa = 10.68HH91 pKa = 6.85PYY93 pKa = 9.68IEE95 pKa = 5.0KK96 pKa = 10.28IRR98 pKa = 11.84GLRR101 pKa = 11.84NMIKK105 pKa = 10.22YY106 pKa = 7.58CTKK109 pKa = 10.72EE110 pKa = 3.88DD111 pKa = 4.03TEE113 pKa = 4.27PLANFDD119 pKa = 3.48WHH121 pKa = 6.99PSTEE125 pKa = 4.21NKK127 pKa = 10.37DD128 pKa = 3.66KK129 pKa = 11.56GIDD132 pKa = 3.23WEE134 pKa = 4.45EE135 pKa = 4.37IYY137 pKa = 10.99SVEE140 pKa = 4.18YY141 pKa = 10.07PSAQAFLDD149 pKa = 3.92TVMARR154 pKa = 11.84YY155 pKa = 8.52PSYY158 pKa = 9.96FTGHH162 pKa = 6.86YY163 pKa = 9.83IPLRR167 pKa = 11.84QLAYY171 pKa = 10.47DD172 pKa = 3.72RR173 pKa = 11.84YY174 pKa = 8.52EE175 pKa = 4.53KK176 pKa = 10.78NVEE179 pKa = 4.02EE180 pKa = 4.23YY181 pKa = 10.36RR182 pKa = 11.84PKK184 pKa = 10.67YY185 pKa = 10.81LNFEE189 pKa = 4.23NVPQILNSWVDD200 pKa = 3.64CNLFNDD206 pKa = 3.79KK207 pKa = 10.13EE208 pKa = 4.35RR209 pKa = 11.84PYY211 pKa = 10.52SLIMIGPSRR220 pKa = 11.84TGKK223 pKa = 8.12TEE225 pKa = 3.5WARR228 pKa = 11.84SIGRR232 pKa = 11.84HH233 pKa = 4.97MYY235 pKa = 9.09FCNYY239 pKa = 9.5FNLDD243 pKa = 3.38LWDD246 pKa = 3.97EE247 pKa = 4.39DD248 pKa = 3.25ALYY251 pKa = 10.92AVFDD255 pKa = 5.74DD256 pKa = 4.68MDD258 pKa = 5.5CDD260 pKa = 3.85PDD262 pKa = 4.32IPFEE266 pKa = 4.03KK267 pKa = 10.41CFRR270 pKa = 11.84SWKK273 pKa = 10.55AFFGAQKK280 pKa = 10.49EE281 pKa = 4.23FTVTDD286 pKa = 3.56KK287 pKa = 11.36YY288 pKa = 11.14KK289 pKa = 10.7RR290 pKa = 11.84KK291 pKa = 9.97QKK293 pKa = 10.55VKK295 pKa = 8.81WGKK298 pKa = 9.06PIIWISNNEE307 pKa = 4.09FNCKK311 pKa = 9.79SSTLDD316 pKa = 3.55YY317 pKa = 10.58IRR319 pKa = 11.84KK320 pKa = 9.14NSVRR324 pKa = 11.84VNVYY328 pKa = 10.35SKK330 pKa = 11.11LYY332 pKa = 10.34
MM1 pKa = 7.37SQFQFNHH8 pKa = 5.38IQLGLTYY15 pKa = 9.3PQARR19 pKa = 11.84GLTKK23 pKa = 10.26EE24 pKa = 3.91LLRR27 pKa = 11.84IYY29 pKa = 10.69LEE31 pKa = 3.35NWYY34 pKa = 10.08FNNGTCEE41 pKa = 3.96VRR43 pKa = 11.84VTKK46 pKa = 10.46YY47 pKa = 10.66LIAEE51 pKa = 4.34EE52 pKa = 4.04KK53 pKa = 10.83HH54 pKa = 6.26EE55 pKa = 4.74DD56 pKa = 4.0GEE58 pKa = 4.63PHH60 pKa = 5.36FHH62 pKa = 6.74VYY64 pKa = 10.6LRR66 pKa = 11.84FDD68 pKa = 3.19NKK70 pKa = 9.99LRR72 pKa = 11.84ARR74 pKa = 11.84EE75 pKa = 3.89TRR77 pKa = 11.84LFDD80 pKa = 3.76IFFEE84 pKa = 4.69STGKK88 pKa = 10.53LYY90 pKa = 10.68HH91 pKa = 6.85PYY93 pKa = 9.68IEE95 pKa = 5.0KK96 pKa = 10.28IRR98 pKa = 11.84GLRR101 pKa = 11.84NMIKK105 pKa = 10.22YY106 pKa = 7.58CTKK109 pKa = 10.72EE110 pKa = 3.88DD111 pKa = 4.03TEE113 pKa = 4.27PLANFDD119 pKa = 3.48WHH121 pKa = 6.99PSTEE125 pKa = 4.21NKK127 pKa = 10.37DD128 pKa = 3.66KK129 pKa = 11.56GIDD132 pKa = 3.23WEE134 pKa = 4.45EE135 pKa = 4.37IYY137 pKa = 10.99SVEE140 pKa = 4.18YY141 pKa = 10.07PSAQAFLDD149 pKa = 3.92TVMARR154 pKa = 11.84YY155 pKa = 8.52PSYY158 pKa = 9.96FTGHH162 pKa = 6.86YY163 pKa = 9.83IPLRR167 pKa = 11.84QLAYY171 pKa = 10.47DD172 pKa = 3.72RR173 pKa = 11.84YY174 pKa = 8.52EE175 pKa = 4.53KK176 pKa = 10.78NVEE179 pKa = 4.02EE180 pKa = 4.23YY181 pKa = 10.36RR182 pKa = 11.84PKK184 pKa = 10.67YY185 pKa = 10.81LNFEE189 pKa = 4.23NVPQILNSWVDD200 pKa = 3.64CNLFNDD206 pKa = 3.79KK207 pKa = 10.13EE208 pKa = 4.35RR209 pKa = 11.84PYY211 pKa = 10.52SLIMIGPSRR220 pKa = 11.84TGKK223 pKa = 8.12TEE225 pKa = 3.5WARR228 pKa = 11.84SIGRR232 pKa = 11.84HH233 pKa = 4.97MYY235 pKa = 9.09FCNYY239 pKa = 9.5FNLDD243 pKa = 3.38LWDD246 pKa = 3.97EE247 pKa = 4.39DD248 pKa = 3.25ALYY251 pKa = 10.92AVFDD255 pKa = 5.74DD256 pKa = 4.68MDD258 pKa = 5.5CDD260 pKa = 3.85PDD262 pKa = 4.32IPFEE266 pKa = 4.03KK267 pKa = 10.41CFRR270 pKa = 11.84SWKK273 pKa = 10.55AFFGAQKK280 pKa = 10.49EE281 pKa = 4.23FTVTDD286 pKa = 3.56KK287 pKa = 11.36YY288 pKa = 11.14KK289 pKa = 10.7RR290 pKa = 11.84KK291 pKa = 9.97QKK293 pKa = 10.55VKK295 pKa = 8.81WGKK298 pKa = 9.06PIIWISNNEE307 pKa = 4.09FNCKK311 pKa = 9.79SSTLDD316 pKa = 3.55YY317 pKa = 10.58IRR319 pKa = 11.84KK320 pKa = 9.14NSVRR324 pKa = 11.84VNVYY328 pKa = 10.35SKK330 pKa = 11.11LYY332 pKa = 10.34
Molecular weight: 40.05 kDa
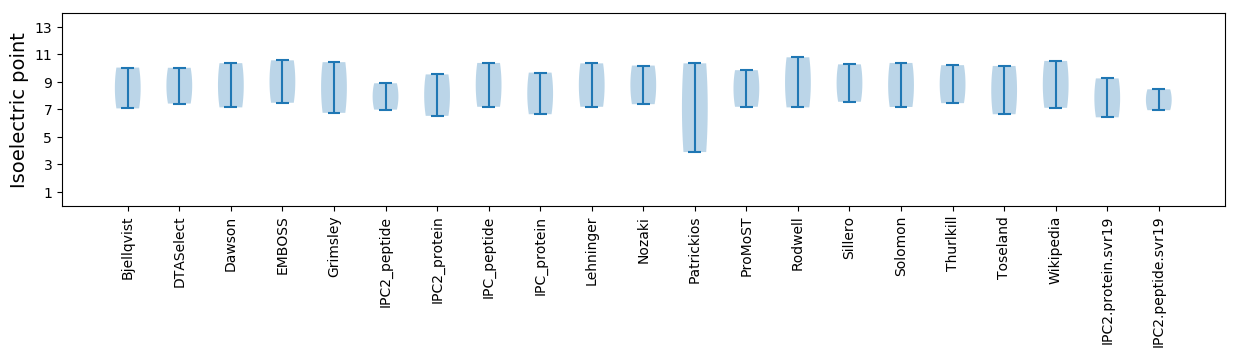
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L0HH91|A0A2L0HH91_9VIRU Replication-associated protein OS=Cattle blood-associated gemycircularvirus OX=2077296 PE=3 SV=1
MM1 pKa = 7.45AGSVKK6 pKa = 10.53FILPTNKK13 pKa = 8.97MPGRR17 pKa = 11.84NNRR20 pKa = 11.84QKK22 pKa = 10.83RR23 pKa = 11.84IATRR27 pKa = 11.84ATNRR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84ITRR37 pKa = 11.84RR38 pKa = 11.84LVRR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 10.23GPDD46 pKa = 2.62IASFMIRR53 pKa = 11.84NLANRR58 pKa = 11.84SNLTSRR64 pKa = 11.84LRR66 pKa = 11.84QRR68 pKa = 11.84LRR70 pKa = 11.84IKK72 pKa = 10.25RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 8.89AKK78 pKa = 9.63YY79 pKa = 8.11VSVRR83 pKa = 11.84ANVAGTTHH91 pKa = 5.81TTSNMIVRR99 pKa = 11.84KK100 pKa = 7.97TPAEE104 pKa = 3.86QKK106 pKa = 10.38FLRR109 pKa = 11.84KK110 pKa = 9.45LFKK113 pKa = 9.92DD114 pKa = 3.38TPNLVKK120 pKa = 10.45HH121 pKa = 5.66VNRR124 pKa = 11.84FAFSWVGASTCSRR137 pKa = 11.84TIWYY141 pKa = 9.79SVAHH145 pKa = 6.17LKK147 pKa = 10.34FNNVYY152 pKa = 10.42DD153 pKa = 4.1YY154 pKa = 11.2LVHH157 pKa = 7.28KK158 pKa = 10.63ISAPNQGVGSAYY170 pKa = 7.4WTTTPTDD177 pKa = 3.25NRR179 pKa = 11.84YY180 pKa = 9.63VSCNPAQFIYY190 pKa = 10.18IGKK193 pKa = 8.6CTFNYY198 pKa = 10.11EE199 pKa = 3.94IYY201 pKa = 10.91NPTNYY206 pKa = 9.89IMTVYY211 pKa = 10.13IYY213 pKa = 11.07DD214 pKa = 4.57LVCKK218 pKa = 9.98HH219 pKa = 6.22DD220 pKa = 3.84TPFEE224 pKa = 3.98ITYY227 pKa = 10.48GSQTDD232 pKa = 3.6VEE234 pKa = 4.64QISRR238 pKa = 11.84SQPEE242 pKa = 3.93CCMIQSSRR250 pKa = 11.84PLTSSAAVDD259 pKa = 3.51AEE261 pKa = 4.01KK262 pKa = 8.66WTVGDD267 pKa = 3.65TTDD270 pKa = 3.97EE271 pKa = 4.36VNNTQQGVGGPAWNAVGMKK290 pKa = 8.21PTDD293 pKa = 3.57YY294 pKa = 11.22FCFNSLWKK302 pKa = 10.8VKK304 pKa = 10.15GIKK307 pKa = 9.98KK308 pKa = 10.08IILPPASSHH317 pKa = 4.99HH318 pKa = 6.22HH319 pKa = 4.46VVVYY323 pKa = 10.38NPKK326 pKa = 10.56KK327 pKa = 10.49KK328 pKa = 8.87ITWGNLVYY336 pKa = 10.24PRR338 pKa = 11.84QDD340 pKa = 2.53ISKK343 pKa = 8.26RR344 pKa = 11.84TGKK347 pKa = 10.68YY348 pKa = 10.36GIGGLTQSTLFGFEE362 pKa = 3.97GQLGFEE368 pKa = 4.79KK369 pKa = 10.82DD370 pKa = 3.22QALDD374 pKa = 3.59NEE376 pKa = 4.89KK377 pKa = 10.97VGTLPGKK384 pKa = 10.21IVVKK388 pKa = 9.82CIRR391 pKa = 11.84KK392 pKa = 8.48INCYY396 pKa = 9.76NFPITSSTVISKK408 pKa = 10.89NNLYY412 pKa = 10.22TNLSNPEE419 pKa = 3.9IFTDD423 pKa = 3.99LTTRR427 pKa = 11.84PAEE430 pKa = 4.11TAA432 pKa = 3.28
MM1 pKa = 7.45AGSVKK6 pKa = 10.53FILPTNKK13 pKa = 8.97MPGRR17 pKa = 11.84NNRR20 pKa = 11.84QKK22 pKa = 10.83RR23 pKa = 11.84IATRR27 pKa = 11.84ATNRR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84ITRR37 pKa = 11.84RR38 pKa = 11.84LVRR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 10.23GPDD46 pKa = 2.62IASFMIRR53 pKa = 11.84NLANRR58 pKa = 11.84SNLTSRR64 pKa = 11.84LRR66 pKa = 11.84QRR68 pKa = 11.84LRR70 pKa = 11.84IKK72 pKa = 10.25RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 8.89AKK78 pKa = 9.63YY79 pKa = 8.11VSVRR83 pKa = 11.84ANVAGTTHH91 pKa = 5.81TTSNMIVRR99 pKa = 11.84KK100 pKa = 7.97TPAEE104 pKa = 3.86QKK106 pKa = 10.38FLRR109 pKa = 11.84KK110 pKa = 9.45LFKK113 pKa = 9.92DD114 pKa = 3.38TPNLVKK120 pKa = 10.45HH121 pKa = 5.66VNRR124 pKa = 11.84FAFSWVGASTCSRR137 pKa = 11.84TIWYY141 pKa = 9.79SVAHH145 pKa = 6.17LKK147 pKa = 10.34FNNVYY152 pKa = 10.42DD153 pKa = 4.1YY154 pKa = 11.2LVHH157 pKa = 7.28KK158 pKa = 10.63ISAPNQGVGSAYY170 pKa = 7.4WTTTPTDD177 pKa = 3.25NRR179 pKa = 11.84YY180 pKa = 9.63VSCNPAQFIYY190 pKa = 10.18IGKK193 pKa = 8.6CTFNYY198 pKa = 10.11EE199 pKa = 3.94IYY201 pKa = 10.91NPTNYY206 pKa = 9.89IMTVYY211 pKa = 10.13IYY213 pKa = 11.07DD214 pKa = 4.57LVCKK218 pKa = 9.98HH219 pKa = 6.22DD220 pKa = 3.84TPFEE224 pKa = 3.98ITYY227 pKa = 10.48GSQTDD232 pKa = 3.6VEE234 pKa = 4.64QISRR238 pKa = 11.84SQPEE242 pKa = 3.93CCMIQSSRR250 pKa = 11.84PLTSSAAVDD259 pKa = 3.51AEE261 pKa = 4.01KK262 pKa = 8.66WTVGDD267 pKa = 3.65TTDD270 pKa = 3.97EE271 pKa = 4.36VNNTQQGVGGPAWNAVGMKK290 pKa = 8.21PTDD293 pKa = 3.57YY294 pKa = 11.22FCFNSLWKK302 pKa = 10.8VKK304 pKa = 10.15GIKK307 pKa = 9.98KK308 pKa = 10.08IILPPASSHH317 pKa = 4.99HH318 pKa = 6.22HH319 pKa = 4.46VVVYY323 pKa = 10.38NPKK326 pKa = 10.56KK327 pKa = 10.49KK328 pKa = 8.87ITWGNLVYY336 pKa = 10.24PRR338 pKa = 11.84QDD340 pKa = 2.53ISKK343 pKa = 8.26RR344 pKa = 11.84TGKK347 pKa = 10.68YY348 pKa = 10.36GIGGLTQSTLFGFEE362 pKa = 3.97GQLGFEE368 pKa = 4.79KK369 pKa = 10.82DD370 pKa = 3.22QALDD374 pKa = 3.59NEE376 pKa = 4.89KK377 pKa = 10.97VGTLPGKK384 pKa = 10.21IVVKK388 pKa = 9.82CIRR391 pKa = 11.84KK392 pKa = 8.48INCYY396 pKa = 9.76NFPITSSTVISKK408 pKa = 10.89NNLYY412 pKa = 10.22TNLSNPEE419 pKa = 3.9IFTDD423 pKa = 3.99LTTRR427 pKa = 11.84PAEE430 pKa = 4.11TAA432 pKa = 3.28
Molecular weight: 49.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
764 |
332 |
432 |
382.0 |
44.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.843 ± 0.592 | 2.094 ± 0.009 |
4.712 ± 1.031 | 4.974 ± 1.826 |
5.105 ± 0.972 | 4.974 ± 0.676 |
2.094 ± 0.202 | 6.283 ± 0.358 |
7.592 ± 0.153 | 6.414 ± 0.713 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.702 ± 0.068 | 6.937 ± 0.391 |
4.843 ± 0.208 | 3.141 ± 0.275 |
6.937 ± 0.391 | 6.021 ± 0.768 |
7.461 ± 1.688 | 5.89 ± 1.069 |
2.094 ± 0.394 | 5.89 ± 1.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
